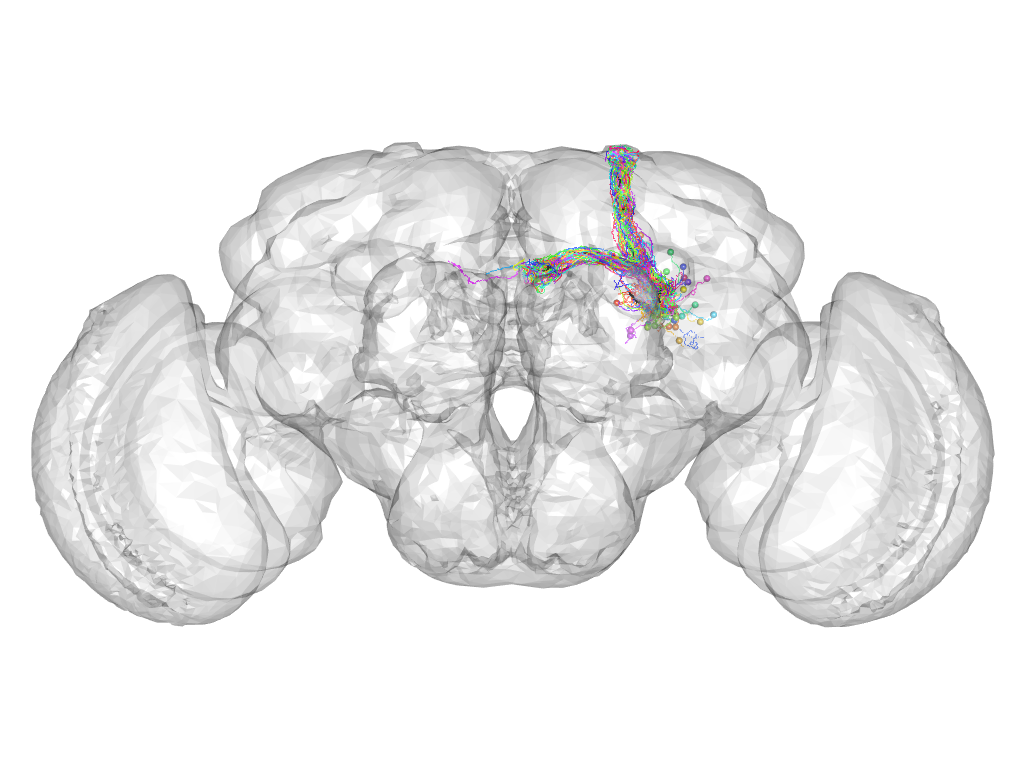
Cluster 311
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-400181), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 79 | fru-M-300247 | XVI | 0.74 |
| 624 | fru-F-400045 | XVI | 0.64 |
| 96 | fru-M-800049 | XVI | 0.63 |
| 501 | fru-F-000031 | XVI | 0.57 |
| 412 | fru-F-200098 | XVI | 0.57 |
| 979 | VGlut-F-400141 | XVI | 0.56 |
| 41 | fru-M-700086 | XVI | 0.56 |
| 599 | VGlut-F-500640 | XVI | 0.56 |
| 931 | VGlut-F-100079 | XVI | 0.55 |
| 630 | fru-F-500012 | XVI | 0.55 |
| 93 | fru-M-300145 | XVI | 0.53 |
| 111 | fru-M-100078 | XVI | 0.52 |
| 538 | Gad1-F-100010 | XVI | 0.51 |
| 945 | VGlut-F-100136 | XVI | 0.51 |
| 116 | fru-M-400058 | XVI | 0.50 |
Cluster composition
This cluster has 44 neurons. The exemplar of this cluster (fru-F-400181) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-400181 | 4694 | FruMARCM-F001929_seg001 | fru-Gal4 | F | 5580.72 | 1.00 |
| fru-M-200212 | 486 | FruMARCM-M001586_seg001 | fru-Gal4 | M | 3734.23 | 0.67 |
| fru-M-100164 | 534 | FruMARCM-M001652_seg002 | fru-Gal4 | M | 3913.16 | 0.68 |
| fru-M-600089 | 867 | FruMARCM-M002122_seg002 | fru-Gal4 | M | 4094.92 | 0.72 |
| fru-M-100099 | 1037 | FruMARCM-M001220_seg002 | fru-Gal4 | M | 4069.21 | 0.70 |
| fru-M-200141 | 1348 | FruMARCM-M001097_seg001 | fru-Gal4 | M | 3782.80 | 0.64 |
| fru-M-300100 | 1430 | FruMARCM-M000941_seg002 | fru-Gal4 | M | 3904.26 | 0.68 |
| fru-M-400049 | 1680 | FruMARCM-M000835_seg002 | fru-Gal4 | M | 3979.67 | 0.69 |
| fru-M-500029 | 1950 | FruMARCM-M000595_seg001 | fru-Gal4 | M | 3144.02 | 0.63 |
| Cha-F-700193 | 3882 | ChaMARCM-F001546_seg002 | Cha-Gal4 | F | 3732.43 | 0.66 |
| Gad1-F-800077 | 4017 | GadMARCM-F000628_seg002 | Gad1-Gal4 | F | 3543.80 | 0.63 |
| Cha-F-100178 | 4927 | ChaMARCM-F001035_seg002 | Cha-Gal4 | F | 3718.40 | 0.64 |
| Cha-F-300271 | 5036 | ChaMARCM-F001121_seg001 | Cha-Gal4 | F | 3330.52 | 0.60 |
| Cha-F-100222 | 5056 | ChaMARCM-F001133_seg001 | Cha-Gal4 | F | 3853.04 | 0.65 |
| Cha-F-200234 | 5128 | ChaMARCM-F001181_seg002 | Cha-Gal4 | F | 3627.02 | 0.63 |
| Cha-F-200356 | 5243 | ChaMARCM-F001408_seg001 | Cha-Gal4 | F | 3494.37 | 0.65 |
| Cha-F-700143 | 5370 | ChaMARCM-F000738_seg001 | Cha-Gal4 | F | 3380.95 | 0.60 |
| Cha-F-600111 | 5427 | ChaMARCM-F000776_seg001 | Cha-Gal4 | F | 3647.78 | 0.65 |
| fru-F-600069 | 6249 | FruMARCM-F001262_seg001 | fru-Gal4 | F | 3794.40 | 0.62 |
| fru-F-000063 | 6355 | FruMARCM-F001500_seg003 | fru-Gal4 | F | 3996.35 | 0.71 |
| fru-F-600086 | 6391 | FruMARCM-F001526_seg002 | fru-Gal4 | F | 3974.66 | 0.73 |
| Cha-F-500124 | 7090 | ChaMARCM-F000569_seg001 | Cha-Gal4 | F | 3704.23 | 0.64 |
| Cha-F-400131 | 7147 | ChaMARCM-F000610_seg002 | Cha-Gal4 | F | 3472.32 | 0.63 |
| Cha-F-000102 | 7160 | ChaMARCM-F000616_seg003 | Cha-Gal4 | F | 3374.22 | 0.58 |
| fru-F-000026 | 7767 | FruMARCM-F000666_seg001 | fru-Gal4 | F | 3444.98 | 0.58 |
| fru-F-900011 | 7831 | FruMARCM-F000732_seg001 | fru-Gal4 | F | 3862.89 | 0.66 |
| Cha-F-700066 | 8207 | ChaMARCM-F000214_seg001 | Cha-Gal4 | F | 3648.62 | 0.60 |
| Cha-F-400052 | 8262 | ChaMARCM-F000247_seg001 | Cha-Gal4 | F | 3332.08 | 0.60 |
| Cha-F-300101 | 8353 | ChaMARCM-F000302_seg001 | Cha-Gal4 | F | 2590.70 | 0.49 |
| Gad1-F-400051 | 8412 | GadMARCM-F000348_seg002 | Gad1-Gal4 | F | 3760.42 | 0.65 |
| VGlut-F-500561 | 8504 | DvGlutMARCM-F003087_seg001 | VGlut-Gal4 | F | 3651.76 | 0.60 |
| Gad1-F-200117 | 9620 | GadMARCM-F000481_seg001 | Gad1-Gal4 | F | 3570.99 | 0.62 |
| fru-F-400055 | 9802 | FruMARCM-F000362_seg002 | fru-Gal4 | F | 3987.62 | 0.68 |
| fru-F-000023 | 9823 | FruMARCM-F000390_seg001 | fru-Gal4 | F | 3432.31 | 0.61 |
| Gad1-F-800006 | 10604 | GadMARCM-F000114_seg001 | Gad1-Gal4 | F | 4001.48 | 0.68 |
| Gad1-F-000007 | 10661 | GadMARCM-F000173_seg001 | Gad1-Gal4 | F | 3281.67 | 0.53 |
| VGlut-F-500474 | 10950 | DvGlutMARCM-F002846_seg001 | VGlut-Gal4 | F | 3387.28 | 0.51 |
| Gad1-F-200055 | 11230 | GadMARCM-F000212_seg001 | Gad1-Gal4 | F | 3861.29 | 0.67 |
| Gad1-F-300068 | 11247 | GadMARCM-F000227_seg001 | Gad1-Gal4 | F | 3617.22 | 0.60 |
| Gad1-F-000021 | 11264 | GadMARCM-F000242_seg001 | Gad1-Gal4 | F | 3681.55 | 0.67 |
| Gad1-F-600043 | 11384 | GadMARCM-F000337_seg001 | Gad1-Gal4 | F | 3382.33 | 0.60 |
| Cha-F-300011 | 11584 | ChaMARCM-F000018_seg002 | Cha-Gal4 | F | 3471.45 | 0.62 |
| VGlut-F-500102 | 12323 | DvGlutMARCM-F473_seg1 | VGlut-Gal4 | F | 3394.78 | 0.58 |
| VGlut-F-100222 | 13091 | DvGlutMARCM-F2113_seg1 | VGlut-Gal4 | F | 3638.21 | 0.62 |