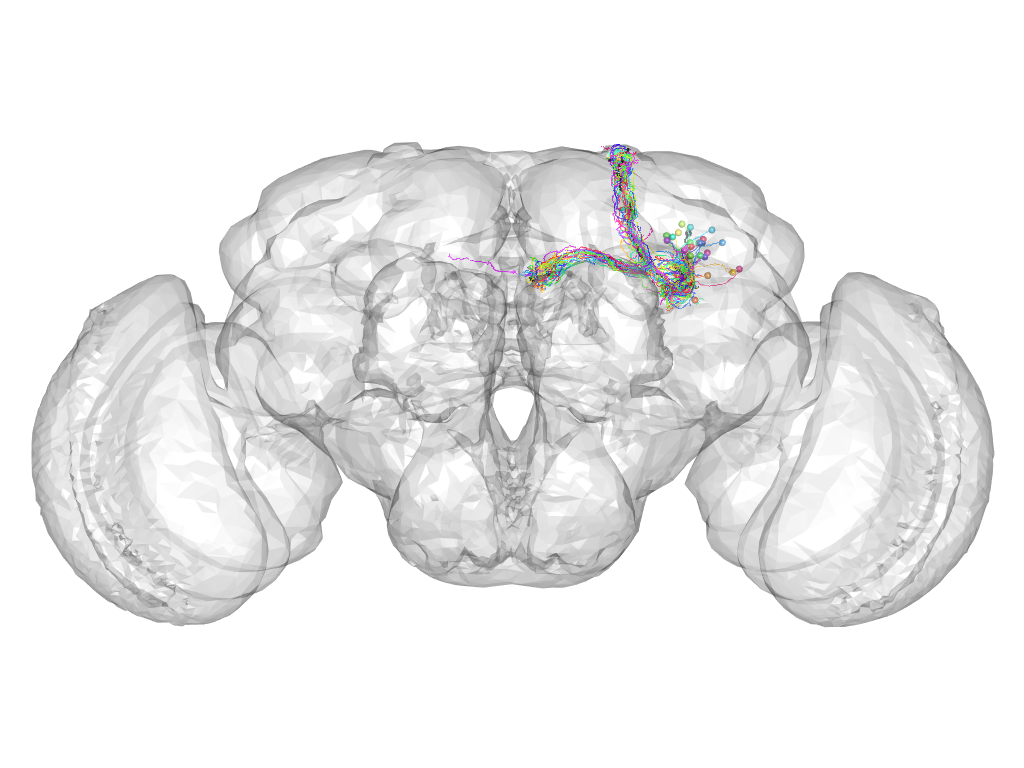
Cluster 538
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Gad1-F-100010), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 630 | fru-F-500012 | XVI | 0.70 |
| 93 | fru-M-300145 | XVI | 0.68 |
| 116 | fru-M-400058 | XVI | 0.67 |
| 501 | fru-F-000031 | XVI | 0.65 |
| 88 | fru-M-500118 | XVI | 0.60 |
Cluster composition
This cluster has 27 neurons. The exemplar of this cluster (Gad1-F-100010) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Gad1-F-100010 | 8399 | GadMARCM-F000050_seg001 | Gad1-Gal4 | F | 4259.57 | 1.00 |
| fru-M-000220 | 410 | FruMARCM-M002649_seg002 | fru-Gal4 | M | 3150.24 | 0.70 |
| fru-M-000110 | 591 | FruMARCM-M001785_seg002 | fru-Gal4 | M | 2833.10 | 0.69 |
| fru-M-200203 | 1295 | FruMARCM-M001487_seg002 | fru-Gal4 | M | 2837.22 | 0.69 |
| fru-M-600014 | 2468 | FruMARCM-M000224_seg003 | fru-Gal4 | M | 2814.28 | 0.63 |
| fru-F-200117 | 4604 | FruMARCM-F001722_seg001 | fru-Gal4 | F | 3111.96 | 0.70 |
| Cha-F-600096 | 7194 | ChaMARCM-F000642_seg001 | Cha-Gal4 | F | 2729.65 | 0.62 |
| Cha-F-000092 | 7262 | ChaMARCM-F000556_seg001 | Cha-Gal4 | F | 3081.45 | 0.72 |
| Cha-F-500096 | 7501 | ChaMARCM-F000363_seg002 | Cha-Gal4 | F | 2910.97 | 0.65 |
| Cha-F-700113 | 7587 | ChaMARCM-F000442_seg001 | Cha-Gal4 | F | 3042.77 | 0.68 |
| Cha-F-600020 | 8188 | ChaMARCM-F000201_seg001 | Cha-Gal4 | F | 2832.97 | 0.62 |
| Cha-F-400043 | 8225 | ChaMARCM-F000226_seg001 | Cha-Gal4 | F | 3001.98 | 0.69 |
| Cha-F-400044 | 8226 | ChaMARCM-F000226_seg002 | Cha-Gal4 | F | 2513.44 | 0.61 |
| Cha-F-000057 | 8348 | ChaMARCM-F000299_seg001 | Cha-Gal4 | F | 2844.65 | 0.65 |
| Gad1-F-000046 | 9540 | GadMARCM-F000422_seg001 | Gad1-Gal4 | F | 2905.72 | 0.68 |
| Gad1-F-600058 | 9558 | GadMARCM-F000434_seg001 | Gad1-Gal4 | F | 2952.06 | 0.66 |
| Gad1-F-500079 | 9645 | GadMARCM-F000498_seg001 | Gad1-Gal4 | F | 3181.34 | 0.71 |
| Gad1-F-500084 | 9701 | GadMARCM-F000545_seg002 | Gad1-Gal4 | F | 2817.36 | 0.63 |
| fru-F-500021 | 9892 | FruMARCM-F000486_seg001 | fru-Gal4 | F | 2930.42 | 0.66 |
| fru-F-600029 | 9953 | FruMARCM-F000546_seg001 | fru-Gal4 | F | 2759.87 | 0.63 |
| Cha-F-500014 | 10113 | ChaMARCM-F000066_seg001 | Cha-Gal4 | F | 2578.19 | 0.63 |
| Gad1-F-300017 | 10703 | GadMARCM-F000036_seg001 | Gad1-Gal4 | F | 2691.53 | 0.64 |
| VGlut-F-600292 | 10777 | DvGlutMARCM-F002539_seg002 | VGlut-Gal4 | F | 3014.03 | 0.61 |
| Gad1-F-400029 | 11257 | GadMARCM-F000236_seg002 | Gad1-Gal4 | F | 2777.26 | 0.63 |
| Gad1-F-200006 | 12618 | GadMARCM-F000022_seg1 | Gad1-Gal4 | F | 2682.84 | 0.64 |
| VGlut-F-900027 | 15066 | DvGlutMARCM-F1403_seg1 | VGlut-Gal4 | F | 2847.79 | 0.68 |
| VGlut-F-200108 | 16210 | DvGlutMARCM-F589_seg1 | VGlut-Gal4 | F | 2800.89 | 0.66 |