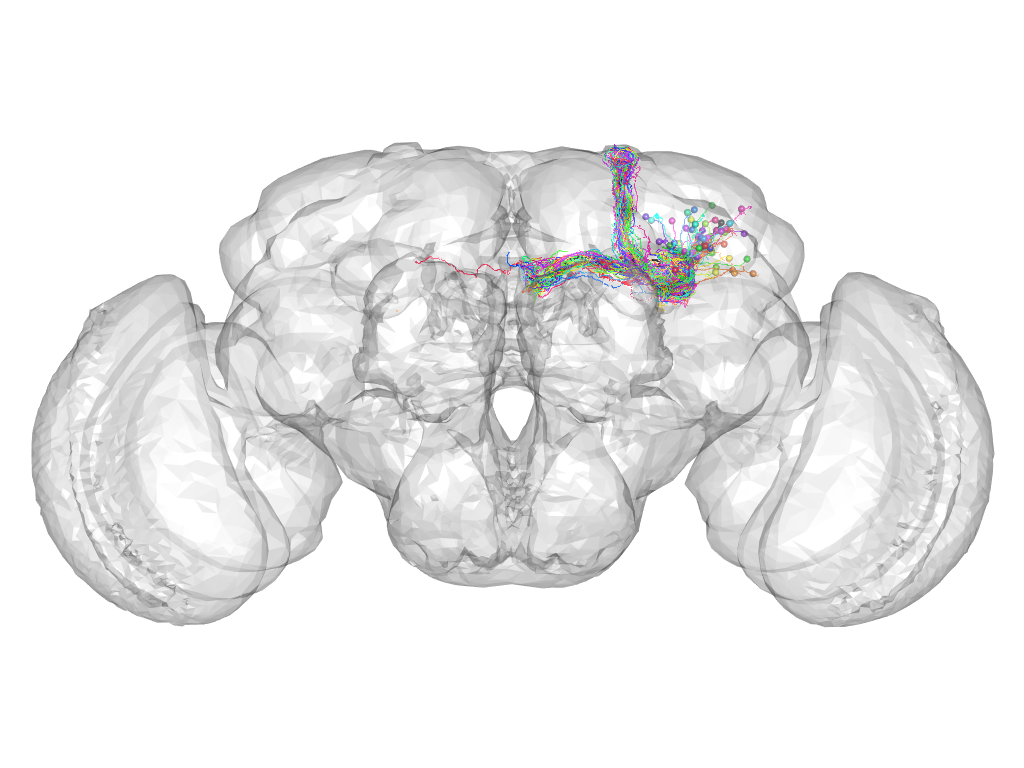
Cluster 116
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-400058), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 501 | fru-F-000031 | XVI | 0.69 |
| 93 | fru-M-300145 | XVI | 0.63 |
| 630 | fru-F-500012 | XVI | 0.61 |
| 538 | Gad1-F-100010 | XVI | 0.60 |
| 88 | fru-M-500118 | XVI | 0.48 |
Cluster composition
This cluster has 60 neurons. The exemplar of this cluster (fru-M-400058) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-400058 | 1689 | FruMARCM-M000842_seg002 | fru-Gal4 | M | 5204.88 | 1.00 |
| fru-M-300252 | 56 | FruMARCM-M002252_seg002 | fru-Gal4 | M | 3683.87 | 0.71 |
| fru-M-200280 | 117 | FruMARCM-M002314_seg002 | fru-Gal4 | M | 3463.97 | 0.68 |
| fru-M-900065 | 174 | FruMARCM-M002391_seg002 | fru-Gal4 | M | 3670.31 | 0.71 |
| fru-M-300172 | 425 | FruMARCM-M001544_seg002 | fru-Gal4 | M | 3424.26 | 0.65 |
| fru-M-800064 | 550 | FruMARCM-M001705_seg001 | fru-Gal4 | M | 3528.88 | 0.68 |
| fru-M-700012 | 951 | FruMARCM-M000151_seg002 | fru-Gal4 | M | 3625.62 | 0.66 |
| fru-M-700075 | 1001 | FruMARCM-M001183_seg001 | fru-Gal4 | M | 3609.22 | 0.71 |
| fru-M-500126 | 1060 | FruMARCM-M001237_seg002 | fru-Gal4 | M | 3639.94 | 0.71 |
| fru-M-200171 | 1109 | FruMARCM-M001291_seg002 | fru-Gal4 | M | 3169.08 | 0.61 |
| fru-M-200191 | 1215 | FruMARCM-M001399_seg002 | fru-Gal4 | M | 3778.53 | 0.71 |
| fru-M-500134 | 1220 | FruMARCM-M001404_seg002 | fru-Gal4 | M | 3361.33 | 0.62 |
| fru-M-400111 | 1347 | FruMARCM-M001096_seg004 | fru-Gal4 | M | 3455.69 | 0.66 |
| fru-M-700062 | 1382 | FruMARCM-M000888_seg001 | fru-Gal4 | M | 3383.33 | 0.65 |
| fru-M-400089 | 1464 | FruMARCM-M001024_seg002 | fru-Gal4 | M | 3569.07 | 0.69 |
| fru-M-300112 | 1502 | FruMARCM-M001053_seg001 | fru-Gal4 | M | 3733.99 | 0.75 |
| fru-M-200132 | 1546 | FruMARCM-M001080_seg001 | fru-Gal4 | M | 3699.67 | 0.71 |
| fru-M-300119 | 1555 | FruMARCM-M001087_seg001 | fru-Gal4 | M | 3398.12 | 0.67 |
| fru-M-300084 | 1638 | FruMARCM-M000777_seg001 | fru-Gal4 | M | 3660.16 | 0.69 |
| fru-M-000014 | 1967 | FruMARCM-M000614_seg001 | fru-Gal4 | M | 3561.59 | 0.72 |
| Gad1-F-900048 | 4026 | GadMARCM-F000635_seg001 | Gad1-Gal4 | F | 3514.09 | 0.69 |
| Cha-F-400217 | 4832 | ChaMARCM-F000958_seg001 | Cha-Gal4 | F | 3372.93 | 0.63 |
| Cha-F-200196 | 4863 | ChaMARCM-F000985_seg001 | Cha-Gal4 | F | 3325.96 | 0.64 |
| Cha-F-000217 | 5022 | ChaMARCM-F001109_seg004 | Cha-Gal4 | F | 3303.54 | 0.64 |
| Cha-F-200267 | 5204 | ChaMARCM-F001237_seg001 | Cha-Gal4 | F | 3442.23 | 0.68 |
| Cha-F-400156 | 5316 | ChaMARCM-F000696_seg001 | Cha-Gal4 | F | 3018.18 | 0.57 |
| Cha-F-300214 | 5449 | ChaMARCM-F000787_seg002 | Cha-Gal4 | F | 3403.02 | 0.67 |
| Cha-F-200159 | 5470 | ChaMARCM-F000800_seg001 | Cha-Gal4 | F | 3212.26 | 0.62 |
| Cha-F-100097 | 5473 | ChaMARCM-F000802_seg002 | Cha-Gal4 | F | 3434.46 | 0.67 |
| Cha-F-600127 | 5591 | ChaMARCM-F000883_seg002 | Cha-Gal4 | F | 3635.57 | 0.69 |
| Cha-F-100115 | 5653 | ChaMARCM-F000925_seg001 | Cha-Gal4 | F | 3404.88 | 0.59 |
| fru-F-200075 | 6215 | FruMARCM-F001138_seg002 | fru-Gal4 | F | 3280.84 | 0.64 |
| fru-F-700154 | 6380 | FruMARCM-F001517_seg002 | fru-Gal4 | F | 3717.17 | 0.71 |
| Cha-F-300123 | 7223 | ChaMARCM-F000390_seg001 | Cha-Gal4 | F | 3002.39 | 0.56 |
| Cha-F-900013 | 7447 | ChaMARCM-F000331_seg001 | Cha-Gal4 | F | 2783.64 | 0.55 |
| Cha-F-800010 | 7515 | ChaMARCM-F000371_seg001 | Cha-Gal4 | F | 3439.76 | 0.69 |
| fru-F-900015 | 7879 | FruMARCM-F000801_seg002 | fru-Gal4 | F | 3558.86 | 0.68 |
| Cha-F-700022 | 8040 | ChaMARCM-F000120_seg003 | Cha-Gal4 | F | 2834.88 | 0.54 |
| Cha-F-300103 | 8355 | ChaMARCM-F000302_seg003 | Cha-Gal4 | F | 3210.98 | 0.63 |
| VGlut-F-800205 | 8908 | DvGlutMARCM-F003482_seg001 | VGlut-Gal4 | F | 3464.83 | 0.66 |
| Gad1-F-600055 | 9476 | GadMARCM-F000373_seg001 | Gad1-Gal4 | F | 3358.40 | 0.65 |
| Gad1-F-400070 | 9480 | GadMARCM-F000374_seg002 | Gad1-Gal4 | F | 3486.79 | 0.68 |
| Gad1-F-100038 | 9547 | GadMARCM-F000427_seg001 | Gad1-Gal4 | F | 3380.85 | 0.64 |
| Gad1-F-300112 | 9579 | GadMARCM-F000450_seg001 | Gad1-Gal4 | F | 3399.34 | 0.64 |
| Gad1-F-800053 | 9670 | GadMARCM-F000523_seg001 | Gad1-Gal4 | F | 3481.37 | 0.68 |
| fru-F-800012 | 9871 | FruMARCM-F000456_seg001 | fru-Gal4 | F | 3428.97 | 0.65 |
| fru-F-500026 | 9907 | FruMARCM-F000493_seg002 | fru-Gal4 | F | 3312.80 | 0.61 |
| Cha-F-500016 | 10133 | ChaMARCM-F000078_seg001 | Cha-Gal4 | F | 3733.19 | 0.70 |
| Gad1-F-600020 | 10594 | GadMARCM-F000109_seg001 | Gad1-Gal4 | F | 3284.66 | 0.60 |
| Gad1-F-900016 | 10627 | GadMARCM-F000128_seg002 | Gad1-Gal4 | F | 3669.06 | 0.71 |
| Gad1-F-200036 | 10654 | GadMARCM-F000150_seg001 | Gad1-Gal4 | F | 3160.66 | 0.60 |
| Gad1-F-000011 | 10665 | GadMARCM-F000174_seg002 | Gad1-Gal4 | F | 3237.69 | 0.64 |
| Gad1-F-400021 | 11241 | GadMARCM-F000223_seg001 | Gad1-Gal4 | F | 3665.96 | 0.69 |
| Gad1-F-500050 | 11282 | GadMARCM-F000254_seg003 | Gad1-Gal4 | F | 3253.64 | 0.57 |
| Gad1-F-500066 | 11354 | GadMARCM-F000314_seg001 | Gad1-Gal4 | F | 3144.61 | 0.57 |
| Gad1-F-400037 | 11366 | GadMARCM-F000322_seg001 | Gad1-Gal4 | F | 3703.06 | 0.72 |
| Gad1-F-200009 | 12621 | GadMARCM-F000026_seg001 | Gad1-Gal4 | F | 3006.37 | 0.55 |
| VGlut-F-100109 | 14786 | DvGlutMARCM-F1135_seg1 | VGlut-Gal4 | F | 2944.14 | 0.55 |
| VGlut-F-700016 | 14978 | DvGlutMARCM-F1317_seg1 | VGlut-Gal4 | F | 3270.36 | 0.55 |
| VGlut-F-500056 | 15900 | DvGlutMARCM-F299-X2_seg2 | VGlut-Gal4 | F | 2924.06 | 0.49 |