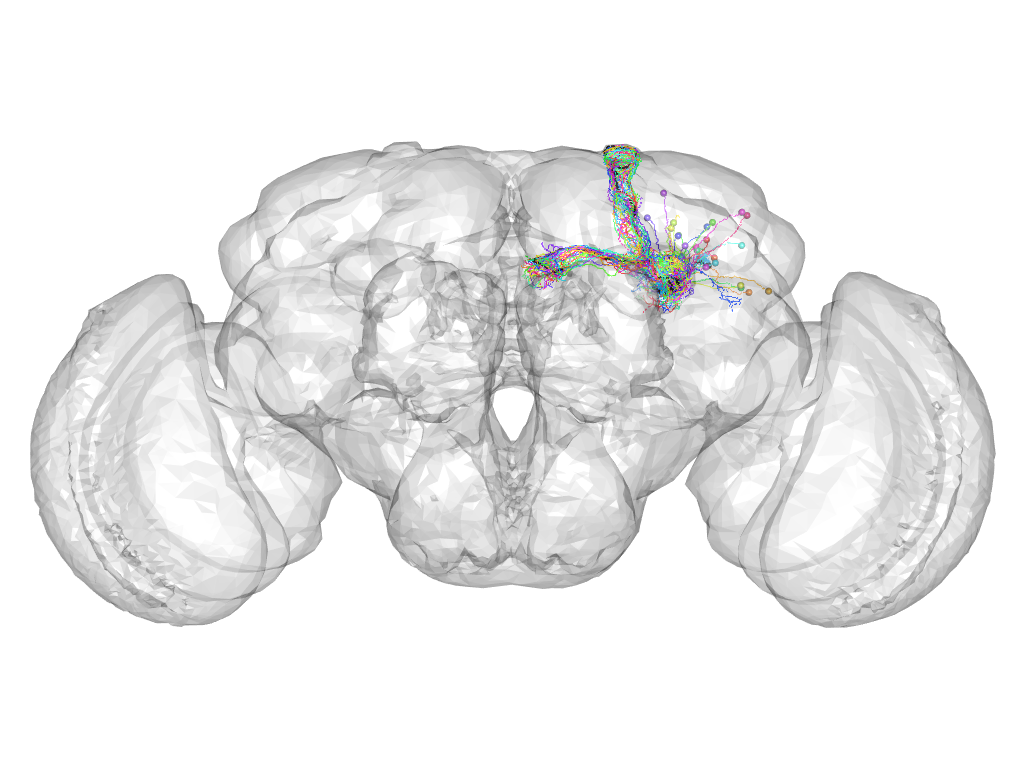
Cluster 88
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-500118), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 630 | fru-F-500012 | XVI | 0.59 |
| 931 | VGlut-F-100079 | XVI | 0.53 |
| 538 | Gad1-F-100010 | XVI | 0.52 |
| 93 | fru-M-300145 | XVI | 0.52 |
| 408 | fru-F-600074 | XVI | 0.50 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (fru-M-500118) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-500118 | 1034 | FruMARCM-M001214_seg001 | fru-Gal4 | M | 7072.71 | 1.00 |
| fru-M-500041 | 1965 | FruMARCM-M000611_seg001 | fru-Gal4 | M | 4444.52 | 0.60 |
| fru-M-400000 | 2442 | FruMARCM-M000206_seg001 | fru-Gal4 | M | 4353.16 | 0.62 |
| fru-M-300029 | 2460 | FruMARCM-M000218_seg002 | fru-Gal4 | M | 4353.73 | 0.60 |
| fru-F-500225 | 4671 | FruMARCM-F001830_seg002 | fru-Gal4 | F | 4124.00 | 0.55 |
| Cha-F-000182 | 4958 | ChaMARCM-F001056_seg001 | Cha-Gal4 | F | 4348.10 | 0.61 |
| Cha-F-000187 | 4966 | ChaMARCM-F001064_seg001 | Cha-Gal4 | F | 4204.67 | 0.60 |
| Cha-F-100200 | 5005 | ChaMARCM-F001097_seg001 | Cha-Gal4 | F | 4138.62 | 0.61 |
| VGlut-F-600736 | 6877 | DvGlutMARCM-F004038_seg001 | VGlut-Gal4 | F | 4303.78 | 0.61 |
| VGlut-F-600749 | 6912 | DvGlutMARCM-F004066_seg001 | VGlut-Gal4 | F | 4000.88 | 0.57 |
| Cha-F-400093 | 7251 | ChaMARCM-F000423_seg001 | Cha-Gal4 | F | 4220.37 | 0.58 |
| Cha-F-700019 | 8029 | ChaMARCM-F000114_seg001 | Cha-Gal4 | F | 3882.00 | 0.54 |
| VGlut-F-500679 | 8942 | DvGlutMARCM-F003508_seg003 | VGlut-Gal4 | F | 4474.78 | 0.62 |
| Gad1-F-100042 | 9601 | GadMARCM-F000468_seg001 | Gad1-Gal4 | F | 4268.16 | 0.60 |
| Gad1-F-600064 | 9650 | GadMARCM-F000503_seg001 | Gad1-Gal4 | F | 4239.08 | 0.59 |
| fru-F-400076 | 9937 | FruMARCM-F000514_seg001 | fru-Gal4 | F | 4636.19 | 0.65 |
| fru-F-400080 | 9973 | FruMARCM-F000564_seg002 | fru-Gal4 | F | 3519.06 | 0.50 |
| Cha-F-400030 | 10132 | ChaMARCM-F000077_seg002 | Cha-Gal4 | F | 2702.94 | 0.46 |
| Gad1-F-500017 | 10501 | GadMARCM-F000153_seg002 | Gad1-Gal4 | F | 4531.53 | 0.54 |
| Gad1-F-600026 | 10522 | GadMARCM-F000163_seg001 | Gad1-Gal4 | F | 4307.98 | 0.63 |
| Gad1-F-000017 | 10671 | GadMARCM-F000178_seg001 | Gad1-Gal4 | F | 4362.79 | 0.61 |
| Gad1-F-300047 | 10696 | GadMARCM-F000195_seg003 | Gad1-Gal4 | F | 2675.25 | 0.45 |
| Gad1-F-100007 | 10720 | GadMARCM-F000048_seg001 | Gad1-Gal4 | F | 4251.32 | 0.57 |
| VGlut-F-000476 | 11088 | DvGlutMARCM-F002961_seg001 | VGlut-Gal4 | F | 4499.39 | 0.64 |
| Gad1-F-200059 | 11234 | GadMARCM-F000216_seg001 | Gad1-Gal4 | F | 4797.33 | 0.64 |
| Gad1-F-200086 | 11332 | GadMARCM-F000301_seg001 | Gad1-Gal4 | F | 4438.80 | 0.64 |
| fru-F-900002 | 11484 | FruMARCM-F000092_seg001 | fru-Gal4 | F | 4560.79 | 0.63 |
| Gad1-F-400001 | 13171 | GadMARCM-F009_seg1 | Gad1-Gal4 | F | 4337.73 | 0.62 |