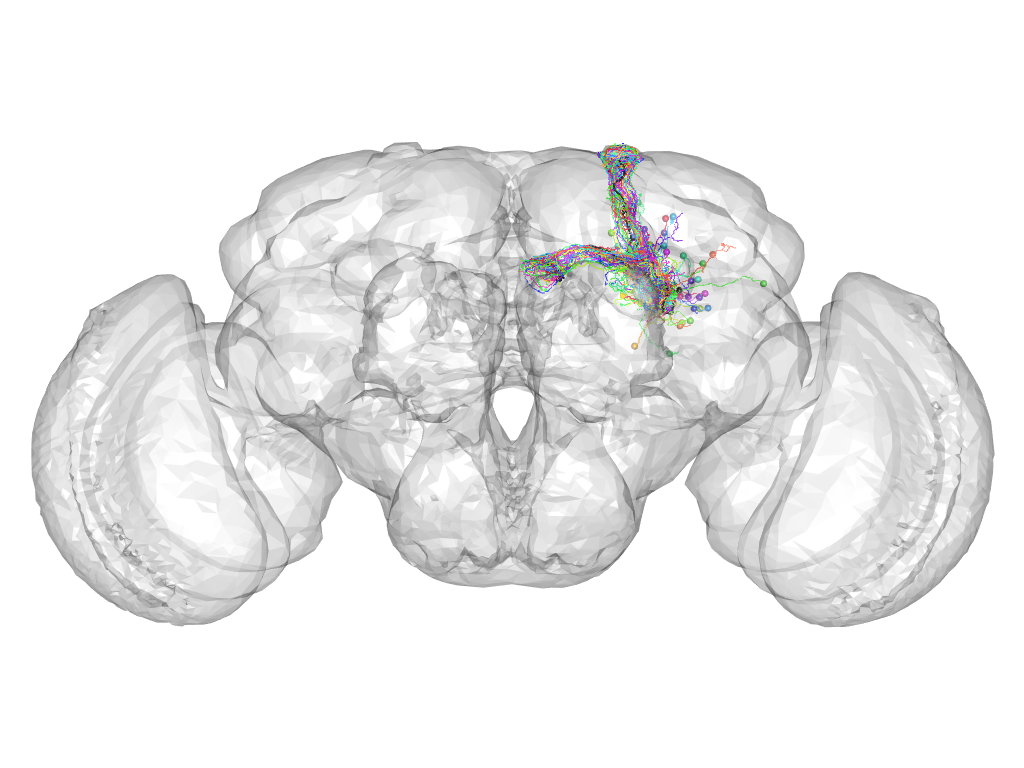
Cluster 931
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-100079), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 88 | fru-M-500118 | XVI | 0.58 |
| 816 | VGlut-F-500310 | XVI | 0.56 |
| 79 | fru-M-300247 | XVI | 0.53 |
| 599 | VGlut-F-500640 | XVI | 0.53 |
| 979 | VGlut-F-400141 | XVI | 0.53 |
| 630 | fru-F-500012 | XVI | 0.51 |
| 101 | fru-M-500143 | XVI | 0.50 |
Cluster composition
This cluster has 39 neurons. The exemplar of this cluster (VGlut-F-100079) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-100079 | 14735 | DvGlutMARCM-F995_seg1 | VGlut-Gal4 | F | 8792.49 | 1.00 |
| Cha-F-100360 | 3928 | ChaMARCM-F001585_seg002 | Cha-Gal4 | F | 5426.83 | 0.64 |
| Cha-F-200223 | 5072 | ChaMARCM-F001141_seg001 | Cha-Gal4 | F | 4871.00 | 0.53 |
| Cha-F-600157 | 5119 | ChaMARCM-F001176_seg001 | Cha-Gal4 | F | 5317.78 | 0.63 |
| Cha-F-400158 | 5333 | ChaMARCM-F000709_seg001 | Cha-Gal4 | F | 5414.91 | 0.63 |
| VGlut-F-800315 | 5911 | DvGlutMARCM-F004577_seg001 | VGlut-Gal4 | F | 5420.12 | 0.63 |
| Trh-F-900012 | 5998 | TPHMARCM-711F_seg3 | Trh-Gal4 | F | 5177.33 | 0.60 |
| fru-F-700121 | 6228 | FruMARCM-F001168_seg001 | fru-Gal4 | F | 5161.61 | 0.61 |
| fru-F-500169 | 6242 | FruMARCM-F001228_seg002 | fru-Gal4 | F | 5230.18 | 0.58 |
| fru-F-500172 | 6268 | FruMARCM-F001302_seg002 | fru-Gal4 | F | 4918.78 | 0.55 |
| fru-F-500179 | 6317 | FruMARCM-F001441_seg002 | fru-Gal4 | F | 5642.15 | 0.64 |
| Cha-F-000104 | 7176 | ChaMARCM-F000629_seg001 | Cha-Gal4 | F | 5087.47 | 0.57 |
| Cha-F-700124 | 7644 | ChaMARCM-F000476_seg001 | Cha-Gal4 | F | 5082.79 | 0.56 |
| fru-F-500106 | 7760 | FruMARCM-F000661_seg002 | fru-Gal4 | F | 4739.79 | 0.50 |
| VGlut-F-200533 | 7938 | DvGlutMARCM-F003773_seg001 | VGlut-Gal4 | F | 4968.36 | 0.54 |
| Cha-F-000037 | 8078 | ChaMARCM-F000144_seg001 | Cha-Gal4 | F | 4841.08 | 0.55 |
| Cha-F-700043 | 8128 | ChaMARCM-F000173_seg001 | Cha-Gal4 | F | 3732.91 | 0.46 |
| Cha-F-200047 | 8237 | ChaMARCM-F000234_seg001 | Cha-Gal4 | F | 4903.61 | 0.52 |
| Cha-F-200054 | 8244 | ChaMARCM-F000238_seg001 | Cha-Gal4 | F | 5019.50 | 0.58 |
| VGlut-F-500567 | 8555 | DvGlutMARCM-F003124_seg001 | VGlut-Gal4 | F | 4980.16 | 0.59 |
| VGlut-F-400763 | 8681 | DvGlutMARCM-F003723_seg001 | VGlut-Gal4 | F | 5672.11 | 0.67 |
| VGlut-F-500652 | 9043 | DvGlutMARCM-F003366_seg001 | VGlut-Gal4 | F | 5404.28 | 0.63 |
| VGlut-F-500660 | 9059 | DvGlutMARCM-F003379_seg002 | VGlut-Gal4 | F | 5291.07 | 0.60 |
| Gad1-F-800027 | 9495 | GadMARCM-F000388_seg001 | Gad1-Gal4 | F | 5306.41 | 0.60 |
| Gad1-F-000051 | 9576 | GadMARCM-F000447_seg001 | Gad1-Gal4 | F | 5474.34 | 0.61 |
| Gad1-F-300117 | 9596 | GadMARCM-F000464_seg002 | Gad1-Gal4 | F | 4958.46 | 0.57 |
| fru-F-500035 | 9916 | FruMARCM-F000499_seg002 | fru-Gal4 | F | 5264.45 | 0.56 |
| Gad1-F-500022 | 10506 | GadMARCM-F000155_seg001 | Gad1-Gal4 | F | 5416.01 | 0.61 |
| Gad1-F-500009 | 10530 | GadMARCM-F000063_seg001 | Gad1-Gal4 | F | 5174.95 | 0.60 |
| Gad1-F-800017 | 11307 | GadMARCM-F000280_seg001 | Gad1-Gal4 | F | 4928.23 | 0.54 |
| Gad1-F-400041 | 11378 | GadMARCM-F000330_seg001 | Gad1-Gal4 | F | 4885.77 | 0.57 |
| Gad1-F-400047 | 11379 | GadMARCM-F000334_seg001 | Gad1-Gal4 | F | 5137.21 | 0.62 |
| Cha-F-300000 | 11570 | ChaMARCM-F000009_seg001 | Cha-Gal4 | F | 4868.68 | 0.58 |
| Trh-F-600099 | 12452 | TPHMARCM-1244F_seg3 | Trh-Gal4 | F | 5300.80 | 0.61 |
| Gad1-F-100002 | 13167 | GadMARCM-F004_seg1 | Gad1-Gal4 | F | 5015.65 | 0.57 |
| Trh-F-700013 | 14225 | TPHMARCM-678F_seg2 | Trh-Gal4 | F | 5335.80 | 0.61 |
| VGlut-F-300146 | 14438 | DvGlutMARCM-F729_seg1 | VGlut-Gal4 | F | 3595.27 | 0.54 |
| VGlut-F-500227 | 15339 | DvGlutMARCM-F1684_seg1 | VGlut-Gal4 | F | 5231.92 | 0.61 |
| VGlut-F-500021 | 15733 | DvGlutMARCM-F144_seg2 | VGlut-Gal4 | F | 5510.32 | 0.63 |