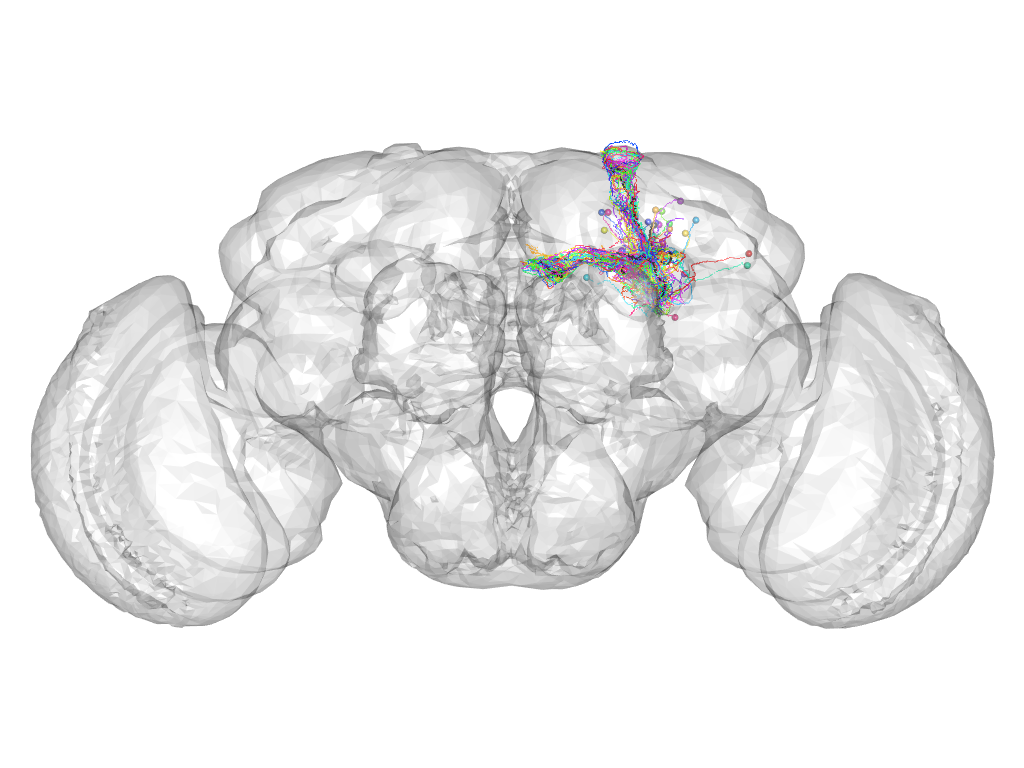
Cluster 816
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-500310), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 931 | VGlut-F-100079 | XVI | 0.61 |
| 599 | VGlut-F-500640 | XVI | 0.57 |
| 88 | fru-M-500118 | XVI | 0.52 |
| 833 | VGlut-F-500352 | XVI | 0.51 |
| 979 | VGlut-F-400141 | XVI | 0.48 |
Cluster composition
This cluster has 30 neurons. The exemplar of this cluster (VGlut-F-500310) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-500310 | 12891 | DvGlutMARCM-F1934_seg1 | VGlut-Gal4 | F | 7368.83 | 1.00 |
| fru-M-500147 | 443 | FruMARCM-M001555_seg002 | fru-Gal4 | M | 4091.49 | 0.54 |
| fru-M-600061 | 1261 | FruMARCM-M001452_seg003 | fru-Gal4 | M | 4643.50 | 0.63 |
| fru-M-500092 | 1445 | FruMARCM-M001014_seg002 | fru-Gal4 | M | 4398.77 | 0.59 |
| Cha-F-200314 | 4257 | ChaMARCM-F001339_seg002 | Cha-Gal4 | F | 3681.19 | 0.52 |
| fru-F-400195 | 4744 | FruMARCM-F002066_seg002 | fru-Gal4 | F | 4072.39 | 0.57 |
| Cha-F-400168 | 5379 | ChaMARCM-F000744_seg001 | Cha-Gal4 | F | 4059.64 | 0.58 |
| VGlut-F-400813 | 6734 | DvGlutMARCM-F003941_seg001 | VGlut-Gal4 | F | 4713.00 | 0.63 |
| fru-F-400113 | 7331 | FruMARCM-F000968_seg001 | fru-Gal4 | F | 4186.00 | 0.57 |
| fru-F-700098 | 7343 | FruMARCM-F000978_seg002 | fru-Gal4 | F | 4586.23 | 0.62 |
| fru-F-500108 | 7762 | FruMARCM-F000662_seg001 | fru-Gal4 | F | 4508.19 | 0.61 |
| VGlut-F-400777 | 7912 | DvGlutMARCM-F003756_seg001 | VGlut-Gal4 | F | 4588.32 | 0.61 |
| Cha-F-600011 | 8047 | ChaMARCM-F000123_seg001 | Cha-Gal4 | F | 4289.64 | 0.60 |
| Cha-F-400063 | 8383 | ChaMARCM-F000323_seg001 | Cha-Gal4 | F | 4422.57 | 0.62 |
| VGlut-F-700294 | 8536 | DvGlutMARCM-F003112_seg002 | VGlut-Gal4 | F | 4459.99 | 0.60 |
| VGlut-F-400764 | 8682 | DvGlutMARCM-F003723_seg002 | VGlut-Gal4 | F | 4771.05 | 0.63 |
| VGlut-F-500678 | 8941 | DvGlutMARCM-F003508_seg002 | VGlut-Gal4 | F | 4403.61 | 0.57 |
| Gad1-F-300114 | 9581 | GadMARCM-F000451_seg001 | Gad1-Gal4 | F | 4567.17 | 0.59 |
| Gad1-F-200113 | 9587 | GadMARCM-F000456_seg001 | Gad1-Gal4 | F | 4653.48 | 0.63 |
| fru-F-500006 | 9759 | FruMARCM-F000271_seg001 | fru-Gal4 | F | 5013.93 | 0.65 |
| fru-F-500072 | 9998 | FruMARCM-F000583_seg002 | fru-Gal4 | F | 4957.26 | 0.67 |
| fru-F-500084 | 10016 | FruMARCM-F000604_seg002 | fru-Gal4 | F | 4157.74 | 0.58 |
| Gad1-F-900015 | 10626 | GadMARCM-F000128_seg001 | Gad1-Gal4 | F | 4659.02 | 0.62 |
| VGlut-F-400609 | 10919 | DvGlutMARCM-F002816_seg001 | VGlut-Gal4 | F | 4198.97 | 0.55 |
| VGlut-F-600231 | 11833 | DvGlutMARCM-F002392_seg001 | VGlut-Gal4 | F | 4243.08 | 0.58 |
| VGlut-F-600378 | 12224 | DvGlutMARCM-F002689_seg002 | VGlut-Gal4 | F | 4242.34 | 0.57 |
| VGlut-F-600103 | 12914 | DvGlutMARCM-F1953_seg2 | VGlut-Gal4 | F | 4339.78 | 0.58 |
| VGlut-F-400506 | 13149 | DvGlutMARCM-F2182_seg1 | VGlut-Gal4 | F | 4042.53 | 0.56 |
| VGlut-F-300297 | 15244 | DvGlutMARCM-F1565_seg1 | VGlut-Gal4 | F | 4455.09 | 0.58 |
| VGlut-F-400102 | 16189 | DvGlutMARCM-F571-X3_seg3 | VGlut-Gal4 | F | 4004.70 | 0.53 |