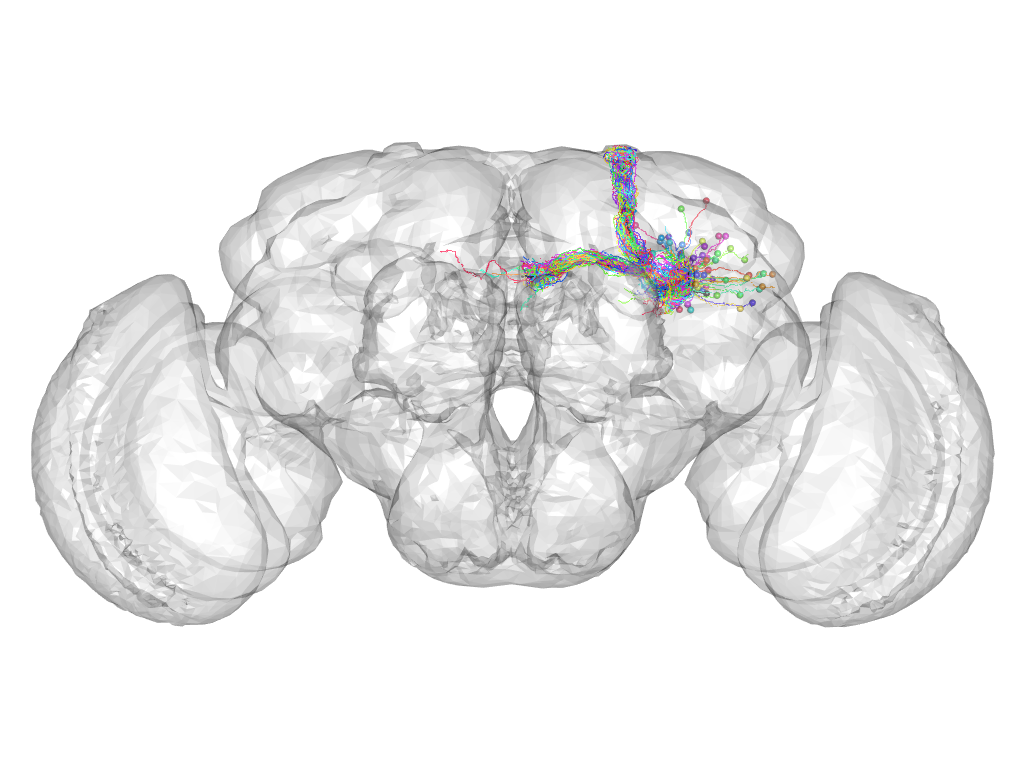
Cluster 93
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-300145), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 501 | fru-F-000031 | XVI | 0.71 |
| 116 | fru-M-400058 | XVI | 0.69 |
| 538 | Gad1-F-100010 | XVI | 0.69 |
| 630 | fru-F-500012 | XVI | 0.64 |
| 88 | fru-M-500118 | XVI | 0.58 |
| 624 | fru-F-400045 | XVI | 0.55 |
| 79 | fru-M-300247 | XVI | 0.54 |
Cluster composition
This cluster has 53 neurons. The exemplar of this cluster (fru-M-300145) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-300145 | 1145 | FruMARCM-M001339_seg001 | fru-Gal4 | M | 4965.70 | 1.00 |
| fru-M-200337 | 13 | FruMARCM-M002591_seg002 | fru-Gal4 | M | 3255.01 | 0.66 |
| fru-M-400210 | 129 | FruMARCM-M002324_seg002 | fru-Gal4 | M | 3211.72 | 0.61 |
| fru-M-300275 | 197 | FruMARCM-M002412_seg003 | fru-Gal4 | M | 3618.74 | 0.72 |
| fru-M-300279 | 213 | FruMARCM-M002429_seg002 | fru-Gal4 | M | 3407.38 | 0.69 |
| fru-M-000079 | 498 | FruMARCM-M001593_seg002 | fru-Gal4 | M | 3469.59 | 0.65 |
| fru-M-500179 | 687 | FruMARCM-M001893_seg001 | fru-Gal4 | M | 3491.90 | 0.67 |
| fru-M-000159 | 869 | FruMARCM-M002123_seg002 | fru-Gal4 | M | 3488.40 | 0.71 |
| fru-M-300125 | 972 | FruMARCM-M001146_seg002 | fru-Gal4 | M | 3450.94 | 0.66 |
| fru-M-200150 | 993 | FruMARCM-M001174_seg002 | fru-Gal4 | M | 3596.94 | 0.69 |
| fru-M-700076 | 1014 | FruMARCM-M001195_seg001 | fru-Gal4 | M | 3247.92 | 0.66 |
| fru-M-100100 | 1038 | FruMARCM-M001220_seg003 | fru-Gal4 | M | 3509.63 | 0.69 |
| fru-M-100112 | 1095 | FruMARCM-M001280_seg003 | fru-Gal4 | M | 3532.32 | 0.71 |
| fru-M-200165 | 1097 | FruMARCM-M001281_seg002 | fru-Gal4 | M | 3401.93 | 0.67 |
| fru-M-100132 | 1174 | FruMARCM-M001358_seg002 | fru-Gal4 | M | 3496.63 | 0.68 |
| fru-M-900037 | 1302 | FruMARCM-M001512_seg002 | fru-Gal4 | M | 3264.45 | 0.59 |
| fru-M-400063 | 1414 | FruMARCM-M000925_seg003 | fru-Gal4 | M | 3165.55 | 0.63 |
| fru-M-300114 | 1504 | FruMARCM-M001055_seg002 | fru-Gal4 | M | 3529.30 | 0.68 |
| fru-M-200126 | 1538 | FruMARCM-M001073_seg003 | fru-Gal4 | M | 3439.54 | 0.67 |
| fru-M-000025 | 1607 | FruMARCM-M000723_seg003 | fru-Gal4 | M | 3480.97 | 0.69 |
| fru-M-300075 | 1610 | FruMARCM-M000726_seg001 | fru-Gal4 | M | 3467.42 | 0.69 |
| fru-M-300082 | 1636 | FruMARCM-M000775_seg001 | fru-Gal4 | M | 3472.50 | 0.66 |
| fru-M-500052 | 1657 | FruMARCM-M000805_seg002 | fru-Gal4 | M | 3376.66 | 0.66 |
| fru-M-100033 | 1872 | FruMARCM-M000415_seg001 | fru-Gal4 | M | 3587.03 | 0.70 |
| fru-M-800012 | 2482 | FruMARCM-M000237_seg001 | fru-Gal4 | M | 3578.63 | 0.67 |
| fru-F-000107 | 4698 | FruMARCM-F001952_seg001 | fru-Gal4 | F | 3597.59 | 0.69 |
| Cha-F-100120 | 4852 | ChaMARCM-F000975_seg002 | Cha-Gal4 | F | 3311.21 | 0.61 |
| Cha-F-100209 | 5043 | ChaMARCM-F001126_seg001 | Cha-Gal4 | F | 2998.51 | 0.61 |
| Cha-F-100220 | 5054 | ChaMARCM-F001131_seg002 | Cha-Gal4 | F | 3290.71 | 0.66 |
| Cha-F-400236 | 5103 | ChaMARCM-F001161_seg001 | Cha-Gal4 | F | 3481.56 | 0.70 |
| Cha-F-200249 | 5156 | ChaMARCM-F001201_seg002 | Cha-Gal4 | F | 3156.78 | 0.60 |
| Cha-F-200251 | 5161 | ChaMARCM-F001204_seg002 | Cha-Gal4 | F | 3070.24 | 0.57 |
| Cha-F-900034 | 5416 | ChaMARCM-F000769_seg001 | Cha-Gal4 | F | 3668.02 | 0.70 |
| VGlut-F-600154 | 6103 | DvGlutMARCM-F2165_seg2 | VGlut-Gal4 | F | 2708.35 | 0.54 |
| Cha-F-000076 | 7240 | ChaMARCM-F000413_seg002 | Cha-Gal4 | F | 2989.74 | 0.58 |
| fru-F-700074 | 7281 | FruMARCM-F000914_seg002 | fru-Gal4 | F | 3551.17 | 0.69 |
| Cha-F-800002 | 7458 | ChaMARCM-F000338_seg002 | Cha-Gal4 | F | 3342.98 | 0.71 |
| fru-F-200067 | 7890 | FruMARCM-F000820_seg001 | fru-Gal4 | F | 3421.16 | 0.65 |
| Cha-F-600017 | 8160 | ChaMARCM-F000187_seg001 | Cha-Gal4 | F | 3046.50 | 0.59 |
| Cha-F-200063 | 8266 | ChaMARCM-F000249_seg001 | Cha-Gal4 | F | 3448.46 | 0.70 |
| Gad1-F-800021 | 8405 | GadMARCM-F000284_seg001 | Gad1-Gal4 | F | 3567.97 | 0.69 |
| VGlut-F-500701 | 8771 | DvGlutMARCM-F003590_seg001 | VGlut-Gal4 | F | 3275.76 | 0.64 |
| VGlut-F-900088 | 8925 | DvGlutMARCM-F003499_seg001 | VGlut-Gal4 | F | 3484.12 | 0.67 |
| Gad1-F-200104 | 9488 | GadMARCM-F000381_seg001 | Gad1-Gal4 | F | 3252.34 | 0.64 |
| Gad1-F-400110 | 9724 | GadMARCM-F000563_seg002 | Gad1-Gal4 | F | 3262.38 | 0.65 |
| fru-F-000014 | 9775 | FruMARCM-F000284_seg001 | fru-Gal4 | F | 3625.31 | 0.68 |
| fru-F-200026 | 9792 | FruMARCM-F000310_seg002 | fru-Gal4 | F | 3454.19 | 0.68 |
| fru-F-900004 | 9794 | FruMARCM-F000336_seg001 | fru-Gal4 | F | 3313.00 | 0.66 |
| Cha-F-400014 | 10083 | ChaMARCM-F000043_seg001 | Cha-Gal4 | F | 3167.09 | 0.62 |
| Gad1-F-100014 | 10675 | GadMARCM-F000181_seg001 | Gad1-Gal4 | F | 3030.21 | 0.57 |
| Gad1-F-200004 | 12616 | GadMARCM-F000020_seg001 | Gad1-Gal4 | F | 3281.68 | 0.63 |
| VGlut-F-400487 | 13032 | DvGlutMARCM-F2056_seg1 | VGlut-Gal4 | F | 2907.75 | 0.49 |
| VGlut-F-200300 | 15317 | DvGlutMARCM-F1656_seg1 | VGlut-Gal4 | F | 3378.19 | 0.64 |