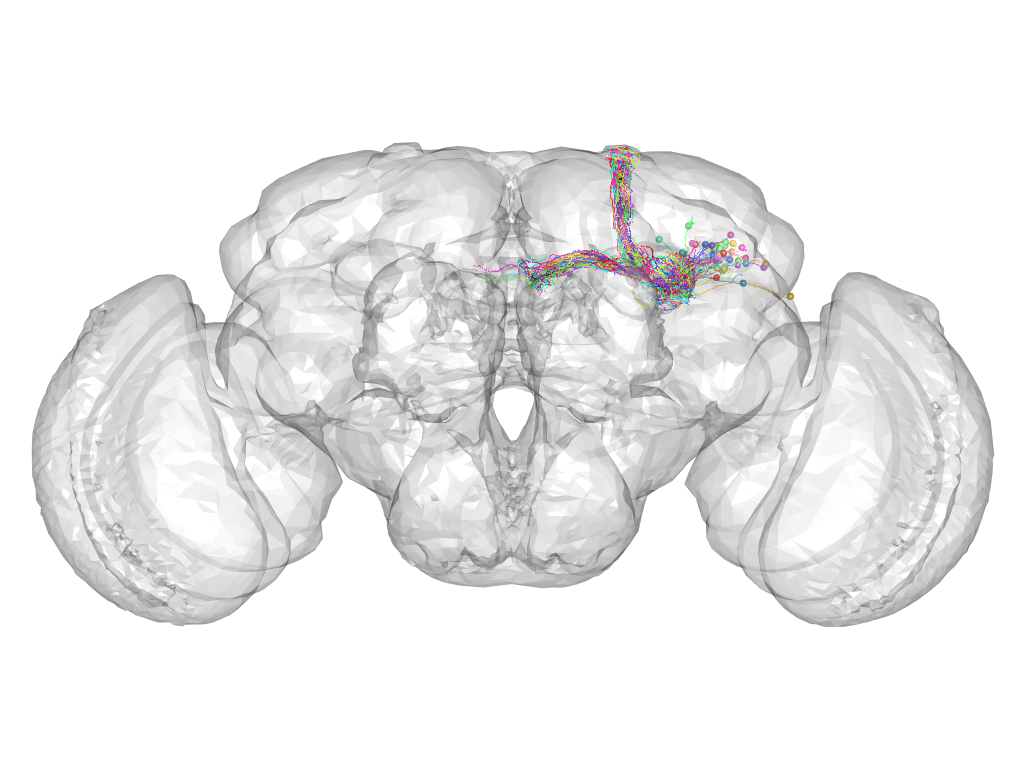
Cluster 501
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-000031), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 116 | fru-M-400058 | XVI | 0.74 |
| 93 | fru-M-300145 | XVI | 0.68 |
| 538 | Gad1-F-100010 | XVI | 0.63 |
| 630 | fru-F-500012 | XVI | 0.59 |
| 624 | fru-F-400045 | XVI | 0.53 |
| 88 | fru-M-500118 | XVI | 0.50 |
Cluster composition
This cluster has 46 neurons. The exemplar of this cluster (fru-F-000031) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-000031 | 7810 | FruMARCM-F000706_seg001 | fru-Gal4 | F | 5227.66 | 1.00 |
| fru-M-100171 | 638 | FruMARCM-M001847_seg002 | fru-Gal4 | M | 3551.23 | 0.68 |
| fru-M-200225 | 664 | FruMARCM-M001872_seg002 | fru-Gal4 | M | 3738.73 | 0.70 |
| fru-M-000132 | 760 | FruMARCM-M001973_seg001 | fru-Gal4 | M | 3839.26 | 0.73 |
| fru-M-400113 | 975 | FruMARCM-M001149_seg001 | fru-Gal4 | M | 3864.08 | 0.73 |
| fru-M-200173 | 1111 | FruMARCM-M001292_seg002 | fru-Gal4 | M | 3571.51 | 0.65 |
| fru-M-900035 | 1244 | FruMARCM-M001423_seg002 | fru-Gal4 | M | 3405.47 | 0.61 |
| fru-M-200194 | 1246 | FruMARCM-M001435_seg002 | fru-Gal4 | M | 3648.11 | 0.68 |
| fru-M-400154 | 1300 | FruMARCM-M001492_seg003 | fru-Gal4 | M | 3684.63 | 0.70 |
| fru-M-500099 | 1332 | FruMARCM-M001090_seg001 | fru-Gal4 | M | 3129.01 | 0.56 |
| fru-M-200142 | 1349 | FruMARCM-M001098_seg001 | fru-Gal4 | M | 3841.26 | 0.72 |
| fru-M-200099 | 1452 | FruMARCM-M001018_seg003 | fru-Gal4 | M | 3587.12 | 0.67 |
| fru-M-400093 | 1469 | FruMARCM-M001029_seg003 | fru-Gal4 | M | 3486.90 | 0.65 |
| fru-M-300109 | 1492 | FruMARCM-M001045_seg002 | fru-Gal4 | M | 3829.18 | 0.71 |
| fru-M-100064 | 1645 | FruMARCM-M000786_seg001 | fru-Gal4 | M | 4007.49 | 0.75 |
| fru-M-700046 | 1665 | FruMARCM-M000813_seg003 | fru-Gal4 | M | 3733.01 | 0.66 |
| fru-M-100023 | 1769 | FruMARCM-M000287_seg002 | fru-Gal4 | M | 3829.08 | 0.73 |
| Cha-F-700195 | 3895 | ChaMARCM-F001556_seg002 | Cha-Gal4 | F | 3306.85 | 0.58 |
| fru-F-000075 | 4496 | FruMARCM-F001600_seg002 | fru-Gal4 | F | 4035.43 | 0.75 |
| fru-F-600119 | 4742 | FruMARCM-F002065_seg002 | fru-Gal4 | F | 3741.95 | 0.72 |
| Cha-F-400167 | 5367 | ChaMARCM-F000735_seg001 | Cha-Gal4 | F | 3370.96 | 0.63 |
| Cha-F-300230 | 5499 | ChaMARCM-F000818_seg002 | Cha-Gal4 | F | 3249.39 | 0.60 |
| Cha-F-500158 | 5629 | ChaMARCM-F000909_seg001 | Cha-Gal4 | F | 3527.99 | 0.66 |
| fru-F-200074 | 6214 | FruMARCM-F001138_seg001 | fru-Gal4 | F | 3420.20 | 0.62 |
| fru-F-700152 | 6338 | FruMARCM-F001491_seg002 | fru-Gal4 | F | 3433.02 | 0.66 |
| fru-F-000067 | 6388 | FruMARCM-F001524_seg002 | fru-Gal4 | F | 3361.47 | 0.61 |
| fru-F-700088 | 7322 | FruMARCM-F000961_seg001 | fru-Gal4 | F | 3947.39 | 0.74 |
| fru-F-300057 | 7381 | FruMARCM-F001007_seg001 | fru-Gal4 | F | 3888.08 | 0.70 |
| fru-F-500144 | 7828 | FruMARCM-F000721_seg001 | fru-Gal4 | F | 3424.44 | 0.65 |
| fru-F-200057 | 7840 | FruMARCM-F000749_seg001 | fru-Gal4 | F | 3995.99 | 0.75 |
| fru-F-900018 | 7903 | FruMARCM-F000847_seg001 | fru-Gal4 | F | 3715.72 | 0.72 |
| Cha-F-900003 | 8090 | ChaMARCM-F000152_seg001 | Cha-Gal4 | F | 3561.98 | 0.67 |
| Cha-F-200036 | 8153 | ChaMARCM-F000183_seg002 | Cha-Gal4 | F | 3368.66 | 0.63 |
| Cha-F-600027 | 8360 | ChaMARCM-F000305_seg001 | Cha-Gal4 | F | 3637.22 | 0.69 |
| VGlut-F-600679 | 8640 | DvGlutMARCM-F003691_seg001 | VGlut-Gal4 | F | 3777.61 | 0.72 |
| Gad1-F-000036 | 9509 | GadMARCM-F000401_seg001 | Gad1-Gal4 | F | 3457.57 | 0.64 |
| Gad1-F-200119 | 9629 | GadMARCM-F000488_seg001 | Gad1-Gal4 | F | 3630.94 | 0.67 |
| fru-F-200043 | 9859 | FruMARCM-F000444_seg001 | fru-Gal4 | F | 3722.74 | 0.70 |
| fru-F-800013 | 9872 | FruMARCM-F000457_seg001 | fru-Gal4 | F | 3030.79 | 0.68 |
| fru-F-400070 | 9886 | FruMARCM-F000480_seg001 | fru-Gal4 | F | 3346.40 | 0.59 |
| fru-F-500055 | 9975 | FruMARCM-F000565_seg002 | fru-Gal4 | F | 3632.82 | 0.68 |
| Cha-F-700013 | 10098 | ChaMARCM-F000053_seg003 | Cha-Gal4 | F | 3375.95 | 0.62 |
| Gad1-F-500021 | 10505 | GadMARCM-F000154_seg002 | Gad1-Gal4 | F | 3173.83 | 0.59 |
| Gad1-F-200054 | 11229 | GadMARCM-F000211_seg001 | Gad1-Gal4 | F | 3531.74 | 0.69 |
| Gad1-F-300069 | 11314 | GadMARCM-F000289_seg001 | Gad1-Gal4 | F | 3753.46 | 0.70 |
| VGlut-F-500320 | 13003 | DvGlutMARCM-F2031_seg2 | VGlut-Gal4 | F | 2995.62 | 0.55 |