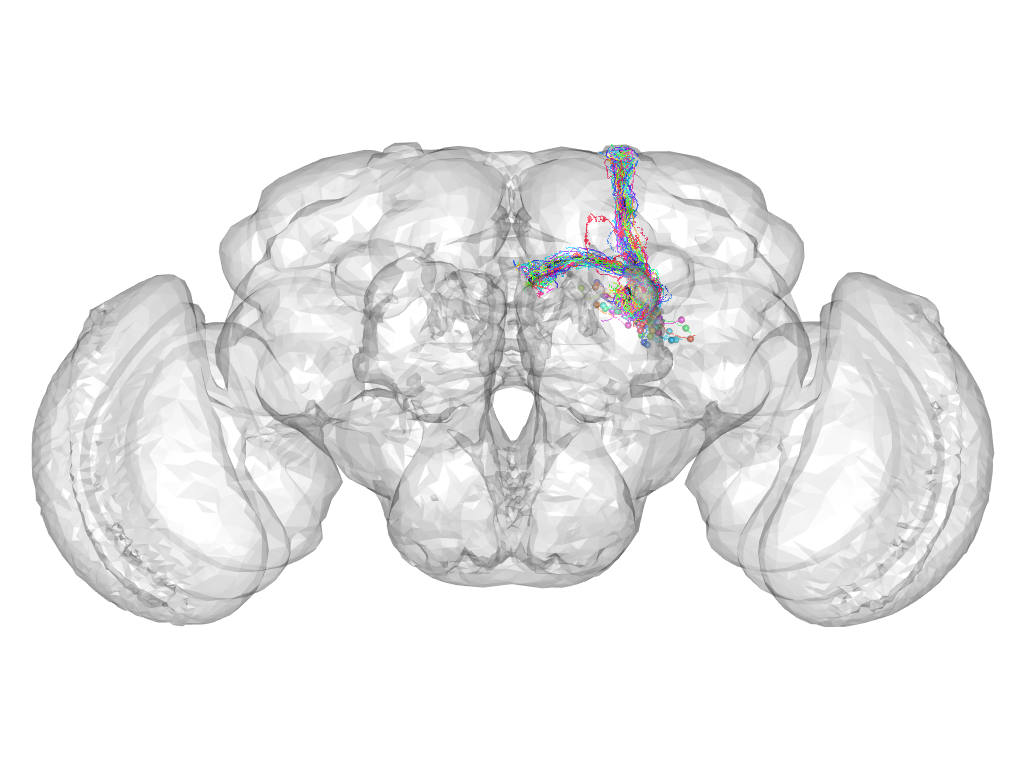
Cluster 96
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-800049), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 412 | fru-F-200098 | XVI | 0.71 |
| 41 | fru-M-700086 | XVI | 0.70 |
| 945 | VGlut-F-100136 | XVI | 0.68 |
| 111 | fru-M-100078 | XVI | 0.68 |
| 311 | fru-F-400181 | XVI | 0.64 |
| 979 | VGlut-F-400141 | XVI | 0.61 |
| 94 | fru-M-600056 | XVI | 0.56 |
| 79 | fru-M-300247 | XVI | 0.54 |
| 101 | fru-M-500143 | XVI | 0.53 |
| 26 | fru-M-200296 | XVI | 0.52 |
| 125 | fru-M-000004 | XVI | 0.52 |
| 419 | fru-F-200021 | XVI | 0.51 |
Cluster composition
This cluster has 41 neurons. The exemplar of this cluster (fru-M-800049) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-800049 | 1224 | FruMARCM-M001408_seg001 | fru-Gal4 | M | 5660.45 | 1.00 |
| fru-M-200261 | 47 | FruMARCM-M002240_seg002 | fru-Gal4 | M | 4299.42 | 0.74 |
| fru-M-100207 | 58 | FruMARCM-M002253_seg002 | fru-Gal4 | M | 3839.32 | 0.68 |
| fru-M-200287 | 161 | FruMARCM-M002382_seg002 | fru-Gal4 | M | 3819.53 | 0.67 |
| fru-M-700169 | 188 | FruMARCM-M002408_seg002 | fru-Gal4 | M | 3580.25 | 0.64 |
| fru-M-300309 | 382 | FruMARCM-M002617_seg003 | fru-Gal4 | M | 4216.99 | 0.72 |
| fru-M-400157 | 508 | FruMARCM-M001611_seg002 | fru-Gal4 | M | 4079.77 | 0.72 |
| fru-M-000136 | 769 | FruMARCM-M001981_seg002 | fru-Gal4 | M | 4044.37 | 0.72 |
| fru-M-300133 | 1046 | FruMARCM-M001226_seg002 | fru-Gal4 | M | 3843.82 | 0.67 |
| fru-M-100129 | 1165 | FruMARCM-M001351_seg002 | fru-Gal4 | M | 4095.14 | 0.72 |
| fru-M-200200 | 1284 | FruMARCM-M001473_seg002 | fru-Gal4 | M | 4112.38 | 0.70 |
| fru-M-500142 | 1309 | FruMARCM-M001527_seg002 | fru-Gal4 | M | 3958.19 | 0.72 |
| fru-M-200123 | 1535 | FruMARCM-M001072_seg001 | fru-Gal4 | M | 3644.78 | 0.66 |
| fru-M-100068 | 1650 | FruMARCM-M000789_seg002 | fru-Gal4 | M | 3779.85 | 0.68 |
| fru-M-900022 | 1659 | FruMARCM-M000807_seg001 | fru-Gal4 | M | 4038.10 | 0.72 |
| fru-M-600018 | 1761 | FruMARCM-M000249_seg003 | fru-Gal4 | M | 3662.42 | 0.63 |
| fru-M-300021 | 2403 | FruMARCM-M000126_seg003 | fru-Gal4 | M | 4023.14 | 0.70 |
| fru-F-400163 | 4515 | FruMARCM-F001625_seg001 | fru-Gal4 | F | 4061.26 | 0.72 |
| Cha-F-200166 | 5534 | ChaMARCM-F000840_seg003 | Cha-Gal4 | F | 3892.88 | 0.69 |
| fru-F-300077 | 6395 | FruMARCM-F001532_seg001 | fru-Gal4 | F | 3694.74 | 0.66 |
| VGlut-F-700528 | 6483 | DvGlutMARCM-F004261_seg002 | VGlut-Gal4 | F | 3018.29 | 0.56 |
| VGlut-F-200580 | 6655 | DvGlutMARCM-F004398_seg001 | VGlut-Gal4 | F | 4129.38 | 0.72 |
| VGlut-F-600714 | 6786 | DvGlutMARCM-F003980_seg002 | VGlut-Gal4 | F | 3536.05 | 0.61 |
| Cha-F-100074 | 7157 | ChaMARCM-F000615_seg002 | Cha-Gal4 | F | 3212.13 | 0.55 |
| Cha-F-100035 | 7550 | ChaMARCM-F000403_seg001 | Cha-Gal4 | F | 3393.42 | 0.62 |
| VGlut-F-700427 | 8018 | DvGlutMARCM-F003833_seg001 | VGlut-Gal4 | F | 3483.52 | 0.58 |
| VGlut-F-300550 | 8020 | DvGlutMARCM-F003835_seg001 | VGlut-Gal4 | F | 3767.37 | 0.67 |
| Cha-F-000034 | 8076 | ChaMARCM-F000142_seg001 | Cha-Gal4 | F | 3270.26 | 0.57 |
| Cha-F-700085 | 8270 | ChaMARCM-F000252_seg002 | Cha-Gal4 | F | 3986.88 | 0.69 |
| Cha-F-500075 | 8295 | ChaMARCM-F000266_seg002 | Cha-Gal4 | F | 3630.64 | 0.61 |
| Cha-F-300099 | 8345 | ChaMARCM-F000297_seg002 | Cha-Gal4 | F | 3798.54 | 0.67 |
| fru-F-200023 | 9731 | FruMARCM-F000241_seg001 | fru-Gal4 | F | 4019.41 | 0.70 |
| fru-F-200030 | 9826 | FruMARCM-F000393_seg001 | fru-Gal4 | F | 3717.13 | 0.68 |
| Cha-F-500012 | 10095 | ChaMARCM-F000052_seg003 | Cha-Gal4 | F | 3791.64 | 0.65 |
| Gad1-F-500011 | 10532 | GadMARCM-F000065_seg001 | Gad1-Gal4 | F | 3742.01 | 0.64 |
| Gad1-F-700001 | 10608 | GadMARCM-F000116_seg002 | Gad1-Gal4 | F | 3372.37 | 0.59 |
| Gad1-F-800011 | 10683 | GadMARCM-F000185_seg003 | Gad1-Gal4 | F | 4017.88 | 0.71 |
| Gad1-F-500046 | 11278 | GadMARCM-F000253_seg001 | Gad1-Gal4 | F | 3667.67 | 0.64 |
| VGlut-F-900048 | 12300 | DvGlutMARCM-F002761_seg001 | VGlut-Gal4 | F | 3853.20 | 0.69 |
| VGlut-F-300101 | 15450 | DvGlutMARCM-F607_seg1 | VGlut-Gal4 | F | 2967.42 | 0.52 |
| VGlut-F-800014 | 15839 | DvGlutMARCM-F246_seg1 | VGlut-Gal4 | F | 4133.70 | 0.61 |