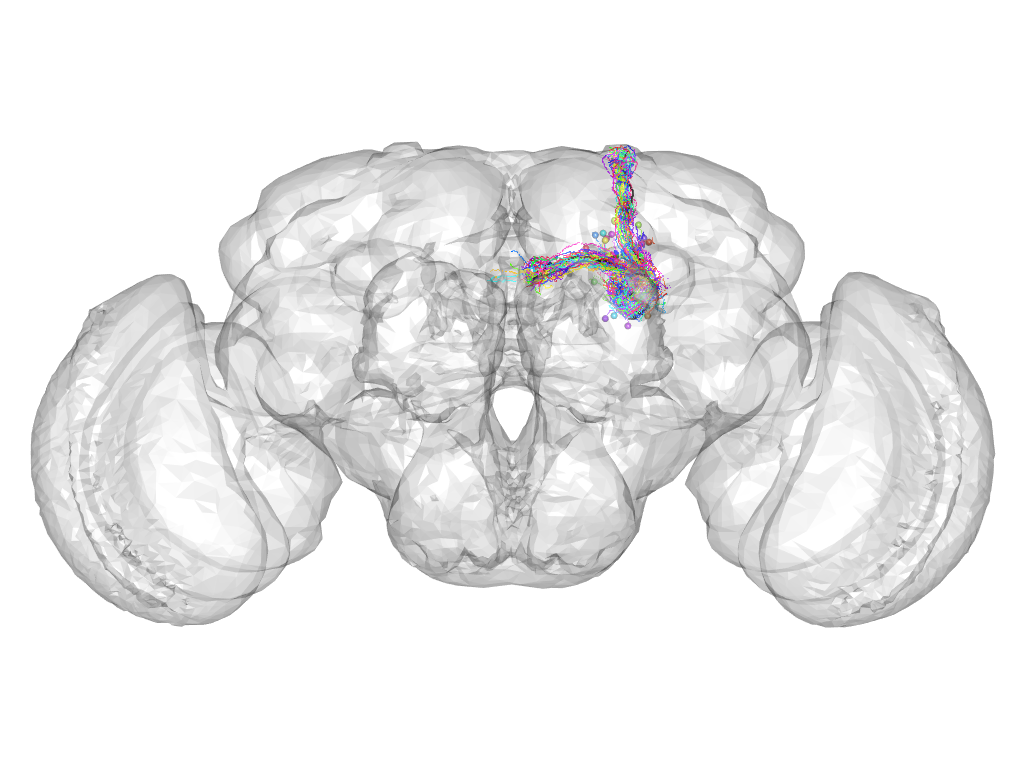
Cluster 41
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-700086), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 96 | fru-M-800049 | XVI | 0.64 |
| 979 | VGlut-F-400141 | XVI | 0.63 |
| 945 | VGlut-F-100136 | XVI | 0.63 |
| 412 | fru-F-200098 | XVI | 0.58 |
| 111 | fru-M-100078 | XVI | 0.57 |
| 26 | fru-M-200296 | XVI | 0.54 |
| 101 | fru-M-500143 | XVI | 0.54 |
| 79 | fru-M-300247 | XVI | 0.53 |
| 94 | fru-M-600056 | XVI | 0.53 |
| 833 | VGlut-F-500352 | XVI | 0.51 |
| 419 | fru-F-200021 | XVI | 0.50 |
| 311 | fru-F-400181 | XVI | 0.50 |
Cluster composition
This cluster has 33 neurons. The exemplar of this cluster (fru-M-700086) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-700086 | 435 | FruMARCM-M001551_seg002 | fru-Gal4 | M | 6070.46 | 1.00 |
| fru-M-600071 | 495 | FruMARCM-M001591_seg002 | fru-Gal4 | M | 3987.47 | 0.62 |
| fru-M-000140 | 789 | FruMARCM-M002003_seg002 | fru-Gal4 | M | 4167.32 | 0.68 |
| fru-M-500212 | 854 | FruMARCM-M002107_seg002 | fru-Gal4 | M | 4077.36 | 0.66 |
| fru-M-200247 | 883 | FruMARCM-M002147_seg002 | fru-Gal4 | M | 4182.20 | 0.67 |
| fru-M-200251 | 905 | FruMARCM-M002170_seg001 | fru-Gal4 | M | 2779.50 | 0.51 |
| fru-M-100104 | 1087 | FruMARCM-M001275_seg001 | fru-Gal4 | M | 3873.69 | 0.60 |
| fru-M-200182 | 1204 | FruMARCM-M001381_seg002 | fru-Gal4 | M | 3497.61 | 0.59 |
| fru-M-200186 | 1208 | FruMARCM-M001382_seg004 | fru-Gal4 | M | 3472.76 | 0.58 |
| fru-M-800041 | 1396 | FruMARCM-M000897_seg001 | fru-Gal4 | M | 3953.88 | 0.65 |
| fru-M-500083 | 1436 | FruMARCM-M001008_seg001 | fru-Gal4 | M | 3909.30 | 0.64 |
| fru-M-200136 | 1550 | FruMARCM-M001083_seg001 | fru-Gal4 | M | 4277.24 | 0.71 |
| fru-M-400045 | 1630 | FruMARCM-M000769_seg001 | fru-Gal4 | M | 4179.53 | 0.68 |
| fru-M-700028 | 1867 | FruMARCM-M000411_seg001 | fru-Gal4 | M | 3780.61 | 0.62 |
| Cha-F-000113 | 5318 | ChaMARCM-F000698_seg001 | Cha-Gal4 | F | 3920.31 | 0.64 |
| Cha-F-800030 | 5410 | ChaMARCM-F000764_seg001 | Cha-Gal4 | F | 4499.73 | 0.73 |
| fru-F-300064 | 6196 | FruMARCM-F001124_seg002 | fru-Gal4 | F | 3988.84 | 0.71 |
| fru-F-300067 | 6204 | FruMARCM-F001130_seg003 | fru-Gal4 | F | 3684.95 | 0.55 |
| fru-F-200104 | 6371 | FruMARCM-F001509_seg002 | fru-Gal4 | F | 4120.85 | 0.70 |
| Cha-F-200120 | 7178 | ChaMARCM-F000631_seg001 | Cha-Gal4 | F | 3614.97 | 0.60 |
| Cha-F-800006 | 7493 | ChaMARCM-F000358_seg001 | Cha-Gal4 | F | 3594.75 | 0.59 |
| VGlut-F-600682 | 8008 | DvGlutMARCM-F003827_seg001 | VGlut-Gal4 | F | 3886.77 | 0.62 |
| Cha-F-300092 | 8321 | ChaMARCM-F000282_seg001 | Cha-Gal4 | F | 3976.93 | 0.62 |
| VGlut-F-800175 | 9079 | DvGlutMARCM-F003394_seg001 | VGlut-Gal4 | F | 4754.33 | 0.74 |
| fru-F-900009 | 9880 | FruMARCM-F000470_seg001 | fru-Gal4 | F | 3724.81 | 0.59 |
| fru-F-500095 | 10034 | FruMARCM-F000624_seg001 | fru-Gal4 | F | 4034.79 | 0.67 |
| Cha-F-200004 | 10071 | ChaMARCM-F000033_seg001 | Cha-Gal4 | F | 3300.98 | 0.50 |
| Gad1-F-400004 | 10536 | GadMARCM-F000068_seg001 | Gad1-Gal4 | F | 3829.80 | 0.63 |
| Gad1-F-300026 | 10547 | GadMARCM-F000075_seg001 | Gad1-Gal4 | F | 3537.60 | 0.56 |
| Gad1-F-900024 | 11297 | GadMARCM-F000270_seg001 | Gad1-Gal4 | F | 3901.82 | 0.64 |
| Gad1-F-400040 | 11377 | GadMARCM-F000329_seg002 | Gad1-Gal4 | F | 3735.62 | 0.60 |
| fru-F-300014 | 11547 | FruMARCM-F000203_seg001 | fru-Gal4 | F | 3299.24 | 0.62 |
| Cha-F-700008 | 11602 | ChaMARCM-F000031_seg002 | Cha-Gal4 | F | 3314.73 | 0.52 |