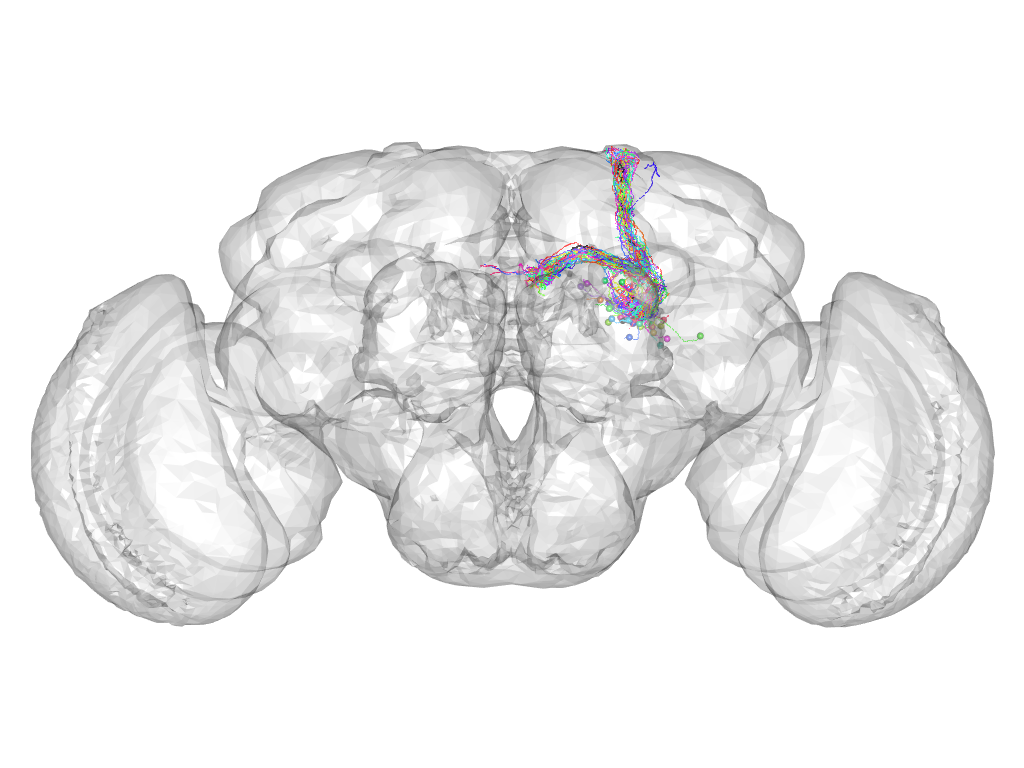
Cluster 412
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-200098), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 96 | fru-M-800049 | XVI | 0.74 |
| 945 | VGlut-F-100136 | XVI | 0.70 |
| 41 | fru-M-700086 | XVI | 0.69 |
| 111 | fru-M-100078 | XVI | 0.67 |
| 311 | fru-F-400181 | XVI | 0.66 |
| 979 | VGlut-F-400141 | XVI | 0.61 |
| 101 | fru-M-500143 | XVI | 0.59 |
| 79 | fru-M-300247 | XVI | 0.57 |
| 94 | fru-M-600056 | XVI | 0.57 |
| 26 | fru-M-200296 | XVI | 0.54 |
| 419 | fru-F-200021 | XVI | 0.53 |
| 599 | VGlut-F-500640 | XVI | 0.52 |
| 833 | VGlut-F-500352 | XVI | 0.52 |
| 125 | fru-M-000004 | XVI | 0.51 |
| 624 | fru-F-400045 | XVI | 0.51 |
Cluster composition
This cluster has 33 neurons. The exemplar of this cluster (fru-F-200098) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-200098 | 6341 | FruMARCM-F001494_seg002 | fru-Gal4 | F | 5478.22 | 1.00 |
| fru-M-500229 | 152 | FruMARCM-M002377_seg002 | fru-Gal4 | M | 3377.61 | 0.56 |
| fru-M-000183 | 224 | FruMARCM-M002438_seg002 | fru-Gal4 | M | 3843.54 | 0.68 |
| fru-M-400178 | 418 | FruMARCM-M001935_seg002 | fru-Gal4 | M | 3819.39 | 0.67 |
| fru-M-000122 | 700 | FruMARCM-M001902_seg002 | fru-Gal4 | M | 4381.52 | 0.77 |
| fru-M-000129 | 744 | FruMARCM-M001958_seg001 | fru-Gal4 | M | 4069.60 | 0.71 |
| fru-M-100109 | 1092 | FruMARCM-M001279_seg001 | fru-Gal4 | M | 4289.38 | 0.76 |
| fru-M-400153 | 1299 | FruMARCM-M001492_seg002 | fru-Gal4 | M | 3850.85 | 0.67 |
| fru-M-100039 | 1878 | FruMARCM-M000419_seg002 | fru-Gal4 | M | 3401.40 | 0.68 |
| fru-M-200006 | 2325 | FruMARCM-M000013_seg001 | fru-Gal4 | M | 3972.57 | 0.69 |
| fru-M-200020 | 2339 | FruMARCM-M000030_seg005 | fru-Gal4 | M | 3418.12 | 0.65 |
| fru-M-100006 | 2356 | FruMARCM-M000049_seg001 | fru-Gal4 | M | 3944.99 | 0.69 |
| Cha-F-100347 | 4421 | ChaMARCM-F001469_seg001 | Cha-Gal4 | F | 3694.36 | 0.63 |
| Cha-F-200209 | 4973 | ChaMARCM-F001069_seg001 | Cha-Gal4 | F | 3750.41 | 0.66 |
| Cha-F-100243 | 5218 | ChaMARCM-F001250_seg002 | Cha-Gal4 | F | 3431.55 | 0.59 |
| Cha-F-200138 | 5312 | ChaMARCM-F000692_seg003 | Cha-Gal4 | F | 3991.60 | 0.71 |
| Cha-F-000114 | 5319 | ChaMARCM-F000698_seg002 | Cha-Gal4 | F | 3615.86 | 0.61 |
| Cha-F-600119 | 5440 | ChaMARCM-F000781_seg002 | Cha-Gal4 | F | 3424.59 | 0.63 |
| Cha-F-900037 | 5507 | ChaMARCM-F000824_seg002 | Cha-Gal4 | F | 3361.63 | 0.65 |
| VGlut-F-400916 | 5818 | DvGlutMARCM-F004511_seg001 | VGlut-Gal4 | F | 3927.77 | 0.70 |
| VGlut-F-300655 | 5924 | DvGlutMARCM-F004584_seg002 | VGlut-Gal4 | F | 3671.68 | 0.62 |
| VGlut-F-800251 | 6811 | DvGlutMARCM-F003999_seg002 | VGlut-Gal4 | F | 3868.43 | 0.67 |
| fru-F-100042 | 7382 | FruMARCM-F001026_seg001 | fru-Gal4 | F | 3730.36 | 0.66 |
| Cha-F-500103 | 7560 | ChaMARCM-F000411_seg001 | Cha-Gal4 | F | 3258.05 | 0.57 |
| Cha-F-300108 | 8363 | ChaMARCM-F000307_seg002 | Cha-Gal4 | F | 3627.88 | 0.65 |
| VGlut-F-500684 | 8947 | DvGlutMARCM-F003512_seg001 | VGlut-Gal4 | F | 3665.82 | 0.61 |
| VGlut-F-300469 | 9325 | DvGlutMARCM-F003163_seg001 | VGlut-Gal4 | F | 3499.90 | 0.59 |
| Gad1-F-400031 | 11268 | GadMARCM-F000246_seg001 | Gad1-Gal4 | F | 3775.29 | 0.65 |
| Gad1-F-100029 | 11275 | GadMARCM-F000250_seg001 | Gad1-Gal4 | F | 3712.49 | 0.64 |
| fru-F-200001 | 11426 | FruMARCM-F000002_seg001 | fru-Gal4 | F | 4090.46 | 0.70 |
| fru-F-800008 | 11534 | FruMARCM-F000192_seg001 | fru-Gal4 | F | 4160.96 | 0.74 |
| VGlut-F-300310 | 15290 | DvGlutMARCM-F1622_seg1 | VGlut-Gal4 | F | 3899.15 | 0.65 |
| VGlut-F-000397 | 15358 | DvGlutMARCM-F1711_seg1 | VGlut-Gal4 | F | 4114.81 | 0.72 |