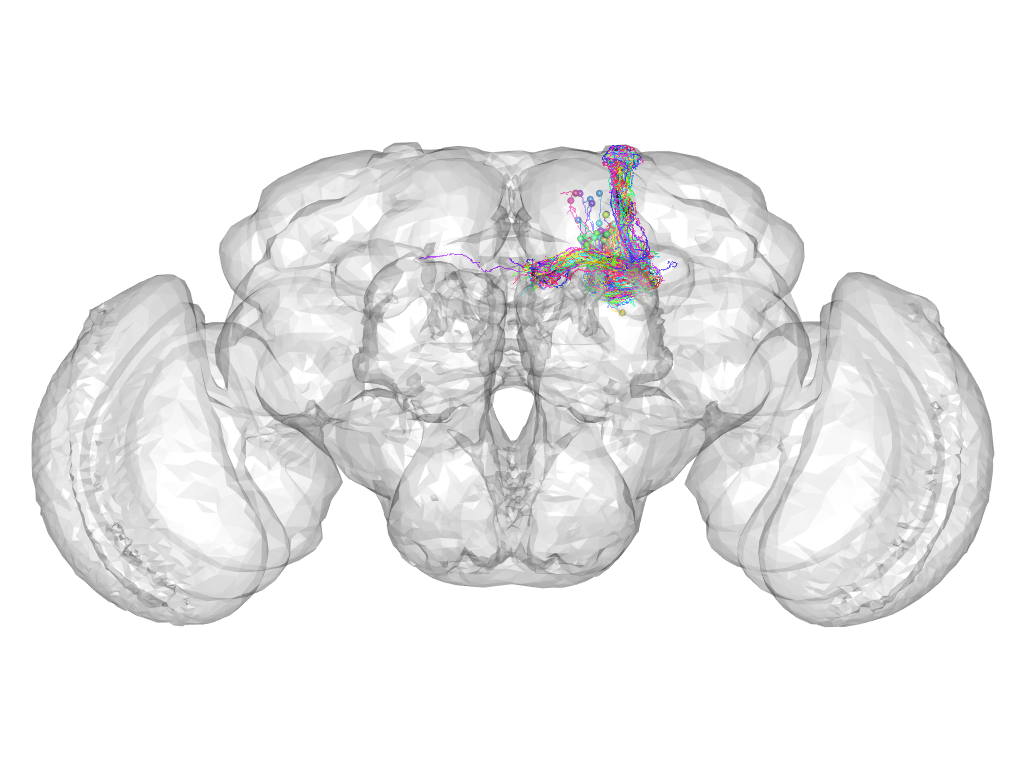
Cluster 26
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-200296), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 419 | fru-F-200021 | XVI | 0.69 |
| 125 | fru-M-000004 | XVI | 0.68 |
| 53 | fru-M-000099 | XVI | 0.64 |
| 522 | Cha-F-300056 | XVI | 0.64 |
| 94 | fru-M-600056 | XVI | 0.63 |
| 533 | Cha-F-900008 | XVI | 0.61 |
| 407 | fru-F-600073 | XVI | 0.58 |
Cluster composition
This cluster has 48 neurons. The exemplar of this cluster (fru-M-200296) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-200296 | 256 | FruMARCM-M002482_seg002 | fru-Gal4 | M | 7459.95 | 1.00 |
| fru-M-500225 | 27 | FruMARCM-M002226_seg002 | fru-Gal4 | M | 5233.01 | 0.69 |
| fru-M-200259 | 42 | FruMARCM-M002237_seg002 | fru-Gal4 | M | 4543.30 | 0.63 |
| fru-M-300274 | 196 | FruMARCM-M002412_seg002 | fru-Gal4 | M | 5246.36 | 0.72 |
| fru-M-200307 | 299 | FruMARCM-M002526_seg002 | fru-Gal4 | M | 5216.50 | 0.71 |
| fru-M-100170 | 637 | FruMARCM-M001847_seg001 | fru-Gal4 | M | 5043.29 | 0.68 |
| fru-M-000137 | 786 | FruMARCM-M002002_seg001 | fru-Gal4 | M | 5380.16 | 0.71 |
| fru-M-600045 | 1028 | FruMARCM-M001208_seg001 | fru-Gal4 | M | 4783.08 | 0.65 |
| fru-M-200163 | 1085 | FruMARCM-M001269_seg001 | fru-Gal4 | M | 4459.14 | 0.64 |
| fru-M-000067 | 1190 | FruMARCM-M001371_seg002 | fru-Gal4 | M | 5213.23 | 0.69 |
| fru-M-200138 | 1554 | FruMARCM-M001086_seg001 | fru-Gal4 | M | 3475.20 | 0.50 |
| fru-M-700029 | 1868 | FruMARCM-M000412_seg001 | fru-Gal4 | M | 5035.13 | 0.68 |
| fru-M-100035 | 1874 | FruMARCM-M000417_seg001 | fru-Gal4 | M | 4283.02 | 0.63 |
| fru-M-100037 | 1876 | FruMARCM-M000418_seg001 | fru-Gal4 | M | 5408.93 | 0.70 |
| fru-M-100047 | 1917 | FruMARCM-M000474_seg001 | fru-Gal4 | M | 4757.69 | 0.68 |
| fru-M-200039 | 2451 | FruMARCM-M000211_seg001 | fru-Gal4 | M | 5276.28 | 0.73 |
| Cha-F-500195 | 3863 | ChaMARCM-F001531_seg002 | Cha-Gal4 | F | 4279.56 | 0.59 |
| Gad1-F-200141 | 4059 | GadMARCM-F000657_seg001 | Gad1-Gal4 | F | 4561.92 | 0.64 |
| Cha-F-100251 | 4164 | ChaMARCM-F001276_seg001 | Cha-Gal4 | F | 4837.39 | 0.65 |
| fru-F-300084 | 4490 | FruMARCM-F001541_seg001 | fru-Gal4 | F | 5267.06 | 0.69 |
| fru-F-500241 | 4732 | FruMARCM-F002058_seg001 | fru-Gal4 | F | 4578.59 | 0.59 |
| Cha-F-400218 | 4833 | ChaMARCM-F000959_seg001 | Cha-Gal4 | F | 4227.31 | 0.58 |
| Cha-F-100188 | 4937 | ChaMARCM-F001042_seg002 | Cha-Gal4 | F | 4257.67 | 0.58 |
| Cha-F-600103 | 5419 | ChaMARCM-F000771_seg001 | Cha-Gal4 | F | 5094.63 | 0.66 |
| Cha-F-600121 | 5478 | ChaMARCM-F000805_seg001 | Cha-Gal4 | F | 4982.36 | 0.69 |
| VGlut-F-000674 | 5869 | DvGlutMARCM-F004553_seg001 | VGlut-Gal4 | F | 5338.18 | 0.71 |
| fru-F-600071 | 6286 | FruMARCM-F001336_seg001 | fru-Gal4 | F | 5146.18 | 0.71 |
| fru-F-200048 | 6406 | FruMARCM-F000463_seg001 | fru-Gal4 | F | 5612.21 | 0.75 |
| Cha-F-600043 | 7217 | ChaMARCM-F000387_seg001 | Cha-Gal4 | F | 4970.81 | 0.66 |
| Cha-F-700096 | 7229 | ChaMARCM-F000404_seg002 | Cha-Gal4 | F | 5037.71 | 0.65 |
| fru-F-800016 | 7769 | FruMARCM-F000667_seg001 | fru-Gal4 | F | 2689.88 | 0.51 |
| fru-F-800025 | 7869 | FruMARCM-F000794_seg001 | fru-Gal4 | F | 3078.77 | 0.57 |
| Cha-F-200059 | 8255 | ChaMARCM-F000242_seg002 | Cha-Gal4 | F | 4127.57 | 0.56 |
| VGlut-F-600511 | 8533 | DvGlutMARCM-F003110_seg002 | VGlut-Gal4 | F | 4760.83 | 0.57 |
| Gad1-F-400071 | 9481 | GadMARCM-F000375_seg001 | Gad1-Gal4 | F | 4753.61 | 0.62 |
| Gad1-F-100057 | 9660 | GadMARCM-F000512_seg001 | Gad1-Gal4 | F | 4732.58 | 0.63 |
| fru-F-700032 | 9881 | FruMARCM-F000472_seg001 | fru-Gal4 | F | 4981.13 | 0.68 |
| fru-F-600024 | 9943 | FruMARCM-F000528_seg001 | fru-Gal4 | F | 4990.28 | 0.57 |
| Gad1-F-600017 | 10591 | GadMARCM-F000107_seg001 | Gad1-Gal4 | F | 4635.96 | 0.65 |
| VGlut-F-400622 | 10841 | DvGlutMARCM-F002853_seg001 | VGlut-Gal4 | F | 4633.61 | 0.63 |
| Gad1-F-500055 | 11286 | GadMARCM-F000257_seg002 | Gad1-Gal4 | F | 4809.75 | 0.62 |
| Gad1-F-400059 | 11405 | GadMARCM-F000352_seg004 | Gad1-Gal4 | F | 4367.12 | 0.57 |
| VGlut-F-600303 | 12031 | DvGlutMARCM-F002551_seg001 | VGlut-Gal4 | F | 4822.13 | 0.60 |
| VGlut-F-400488 | 13033 | DvGlutMARCM-F2057_seg1 | VGlut-Gal4 | F | 5051.49 | 0.66 |
| VGlut-F-500111 | 14528 | DvGlutMARCM-F798_seg1 | VGlut-Gal4 | F | 4578.20 | 0.57 |
| VGlut-F-900006 | 14856 | DvGlutMARCM-F1203_seg1 | VGlut-Gal4 | F | 5413.91 | 0.71 |
| VGlut-F-400149 | 15542 | DvGlutMARCM-F682_seg1 | VGlut-Gal4 | F | 4903.63 | 0.66 |
| VGlut-F-600008 | 15849 | DvGlutMARCM-F256-X2_seg1 | VGlut-Gal4 | F | 4612.36 | 0.61 |