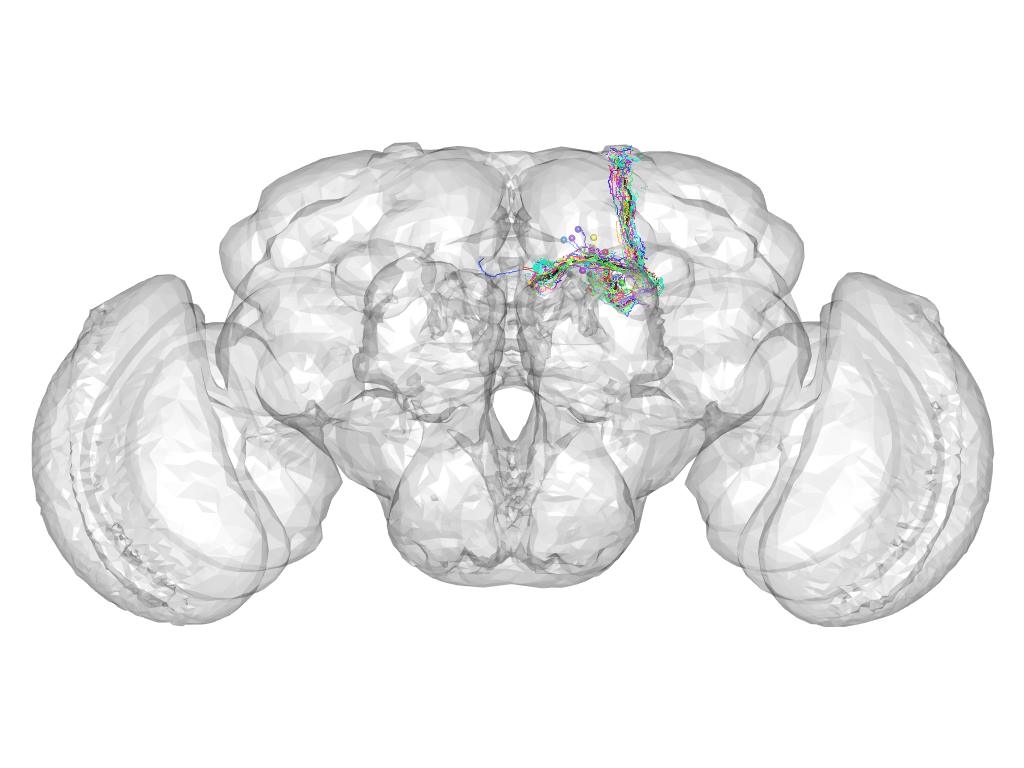
Cluster 94
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-600056), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 26 | fru-M-200296 | XVI | 0.72 |
| 125 | fru-M-000004 | XVI | 0.69 |
| 53 | fru-M-000099 | XVI | 0.69 |
| 419 | fru-F-200021 | XVI | 0.67 |
| 522 | Cha-F-300056 | XVI | 0.61 |
| 945 | VGlut-F-100136 | XVI | 0.60 |
| 41 | fru-M-700086 | XVI | 0.60 |
| 533 | Cha-F-900008 | XVI | 0.56 |
| 111 | fru-M-100078 | XVI | 0.54 |
| 407 | fru-F-600073 | XVI | 0.53 |
| 96 | fru-M-800049 | XVI | 0.51 |
Cluster composition
This cluster has 20 neurons. The exemplar of this cluster (fru-M-600056) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-600056 | 1184 | FruMARCM-M001366_seg002 | fru-Gal4 | M | 6275.47 | 1.00 |
| fru-M-000081 | 518 | FruMARCM-M001631_seg001 | fru-Gal4 | M | 4398.57 | 0.67 |
| fru-M-200250 | 899 | FruMARCM-M002166_seg002 | fru-Gal4 | M | 4635.47 | 0.73 |
| fru-M-300107 | 1485 | FruMARCM-M001041_seg001 | fru-Gal4 | M | 4512.88 | 0.69 |
| fru-M-200068 | 1576 | FruMARCM-M000646_seg002 | fru-Gal4 | M | 4667.24 | 0.73 |
| fru-M-200051 | 1823 | FruMARCM-M000343_seg002 | fru-Gal4 | M | 4440.05 | 0.67 |
| fru-M-100038 | 1877 | FruMARCM-M000419_seg001 | fru-Gal4 | M | 4426.21 | 0.68 |
| fru-M-300025 | 2440 | FruMARCM-M000204_seg001 | fru-Gal4 | M | 4484.54 | 0.71 |
| Cha-F-000288 | 4162 | ChaMARCM-F001273_seg002 | Cha-Gal4 | F | 4139.07 | 0.63 |
| fru-F-700145 | 6334 | FruMARCM-F001484_seg003 | fru-Gal4 | F | 3951.26 | 0.62 |
| Cha-F-300044 | 8139 | ChaMARCM-F000178_seg001 | Cha-Gal4 | F | 4379.68 | 0.61 |
| Cha-F-700072 | 8221 | ChaMARCM-F000223_seg001 | Cha-Gal4 | F | 3691.46 | 0.58 |
| Cha-F-300075 | 8300 | ChaMARCM-F000270_seg001 | Cha-Gal4 | F | 4545.21 | 0.69 |
| VGlut-F-900095 | 8632 | DvGlutMARCM-F003685_seg001 | VGlut-Gal4 | F | 4021.12 | 0.62 |
| Gad1-F-400105 | 9640 | GadMARCM-F000494_seg002 | Gad1-Gal4 | F | 3775.86 | 0.54 |
| Gad1-F-200025 | 10731 | GadMARCM-F000057_seg001 | Gad1-Gal4 | F | 4010.93 | 0.61 |
| Gad1-F-200080 | 11326 | GadMARCM-F000297_seg001 | Gad1-Gal4 | F | 3874.89 | 0.63 |
| fru-F-800006 | 11517 | FruMARCM-F000177_seg001 | fru-Gal4 | F | 3959.26 | 0.66 |
| VGlut-F-600255 | 11859 | DvGlutMARCM-F002407_seg002 | VGlut-Gal4 | F | 4365.11 | 0.65 |
| VGlut-F-800011 | 15772 | DvGlutMARCM-F178_seg2 | VGlut-Gal4 | F | 2866.01 | 0.55 |