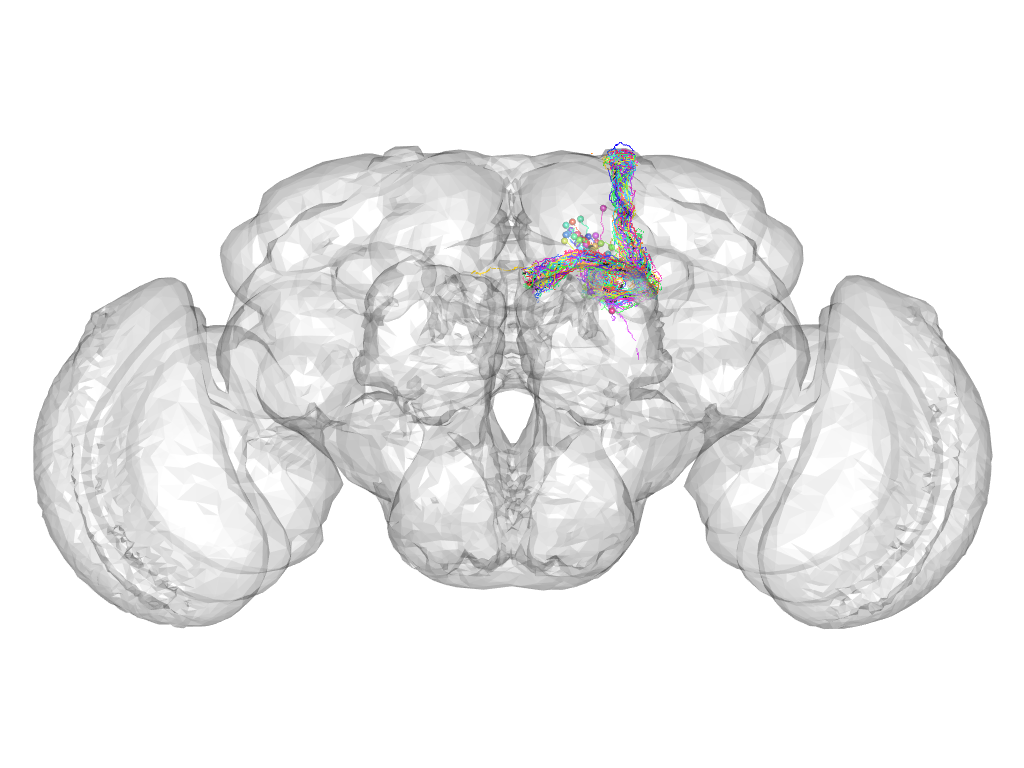
Cluster 419
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-200021), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 26 | fru-M-200296 | XVI | 0.71 |
| 125 | fru-M-000004 | XVI | 0.67 |
| 53 | fru-M-000099 | XVI | 0.61 |
| 94 | fru-M-600056 | XVI | 0.60 |
| 407 | fru-F-600073 | XVI | 0.57 |
| 522 | Cha-F-300056 | XVI | 0.56 |
| 533 | Cha-F-900008 | XVI | 0.55 |
| 945 | VGlut-F-100136 | XVI | 0.53 |
Cluster composition
This cluster has 38 neurons. The exemplar of this cluster (fru-F-200021) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-200021 | 6405 | FruMARCM-F000188_seg001 | fru-Gal4 | F | 5512.39 | 1.00 |
| fru-M-300139 | 1071 | FruMARCM-M001248_seg002 | fru-Gal4 | M | 3493.45 | 0.62 |
| fru-M-900034 | 1243 | FruMARCM-M001423_seg001 | fru-Gal4 | M | 3874.57 | 0.69 |
| fru-M-700066 | 1400 | FruMARCM-M000901_seg001 | fru-Gal4 | M | 3525.25 | 0.64 |
| fru-M-700027 | 1866 | FruMARCM-M000410_seg003 | fru-Gal4 | M | 3645.39 | 0.65 |
| fru-M-400038 | 1887 | FruMARCM-M000423_seg001 | fru-Gal4 | M | 3587.77 | 0.64 |
| fru-F-200109 | 4564 | FruMARCM-F001681_seg001 | fru-Gal4 | F | 3444.29 | 0.60 |
| fru-F-700168 | 4676 | FruMARCM-F001834_seg002 | fru-Gal4 | F | 3729.83 | 0.70 |
| Cha-F-400226 | 4841 | ChaMARCM-F000966_seg001 | Cha-Gal4 | F | 3624.66 | 0.67 |
| Cha-F-100160 | 4905 | ChaMARCM-F001018_seg002 | Cha-Gal4 | F | 3445.18 | 0.64 |
| Cha-F-800035 | 5225 | ChaMARCM-F000807_seg001 | Cha-Gal4 | F | 3590.46 | 0.66 |
| Cha-F-100096 | 5472 | ChaMARCM-F000802_seg001 | Cha-Gal4 | F | 3923.35 | 0.71 |
| Cha-F-100107 | 5552 | ChaMARCM-F000854_seg001 | Cha-Gal4 | F | 3570.05 | 0.66 |
| Cha-F-600094 | 7192 | ChaMARCM-F000640_seg002 | Cha-Gal4 | F | 2712.11 | 0.54 |
| Cha-F-600046 | 7226 | ChaMARCM-F000391_seg001 | Cha-Gal4 | F | 3338.46 | 0.64 |
| Cha-F-200105 | 7724 | ChaMARCM-F000536_seg002 | Cha-Gal4 | F | 3396.11 | 0.60 |
| fru-F-500112 | 7766 | FruMARCM-F000665_seg001 | fru-Gal4 | F | 3283.38 | 0.58 |
| fru-F-800024 | 7839 | FruMARCM-F000738_seg002 | fru-Gal4 | F | 3741.48 | 0.69 |
| fru-F-700062 | 7883 | FruMARCM-F000816_seg001 | fru-Gal4 | F | 3673.04 | 0.65 |
| Cha-F-600014 | 8050 | ChaMARCM-F000124_seg001 | Cha-Gal4 | F | 3258.07 | 0.62 |
| Cha-F-500082 | 8374 | ChaMARCM-F000316_seg001 | Cha-Gal4 | F | 3513.17 | 0.64 |
| Gad1-F-800029 | 9165 | GadMARCM-F000389_seg002 | Gad1-Gal4 | F | 3556.86 | 0.67 |
| Gad1-F-300106 | 9539 | GadMARCM-F000421_seg001 | Gad1-Gal4 | F | 3595.16 | 0.68 |
| fru-F-400066 | 9838 | FruMARCM-F000405_seg002 | fru-Gal4 | F | 3660.87 | 0.66 |
| fru-F-300035 | 9861 | FruMARCM-F000445_seg002 | fru-Gal4 | F | 3680.98 | 0.66 |
| fru-F-700034 | 9894 | FruMARCM-F000487_seg001 | fru-Gal4 | F | 3693.54 | 0.68 |
| fru-F-600025 | 9944 | FruMARCM-F000529_seg001 | fru-Gal4 | F | 3297.10 | 0.62 |
| Cha-F-500034 | 10181 | ChaMARCM-F000109_seg002 | Cha-Gal4 | F | 3530.86 | 0.64 |
| Gad1-F-600024 | 10520 | GadMARCM-F000162_seg001 | Gad1-Gal4 | F | 3340.08 | 0.60 |
| Gad1-F-900010 | 10621 | GadMARCM-F000124_seg002 | Gad1-Gal4 | F | 3886.41 | 0.71 |
| Gad1-F-800020 | 11310 | GadMARCM-F000283_seg001 | Gad1-Gal4 | F | 3214.68 | 0.58 |
| Gad1-F-200076 | 11322 | GadMARCM-F000294_seg001 | Gad1-Gal4 | F | 3560.63 | 0.62 |
| Gad1-F-200094 | 11340 | GadMARCM-F000306_seg002 | Gad1-Gal4 | F | 3680.07 | 0.68 |
| Gad1-F-400048 | 11380 | GadMARCM-F000335_seg001 | Gad1-Gal4 | F | 3270.02 | 0.59 |
| fru-F-500000 | 11497 | FruMARCM-F000162_seg001 | fru-Gal4 | F | 4016.61 | 0.74 |
| fru-F-600004 | 11511 | FruMARCM-F000172_seg001 | fru-Gal4 | F | 3409.40 | 0.63 |
| Gad1-F-100005 | 13170 | GadMARCM-F007_seg1 | Gad1-Gal4 | F | 3344.71 | 0.63 |
| VGlut-F-200120 | 15458 | DvGlutMARCM-F617-X2_seg2 | VGlut-Gal4 | F | 3596.63 | 0.65 |