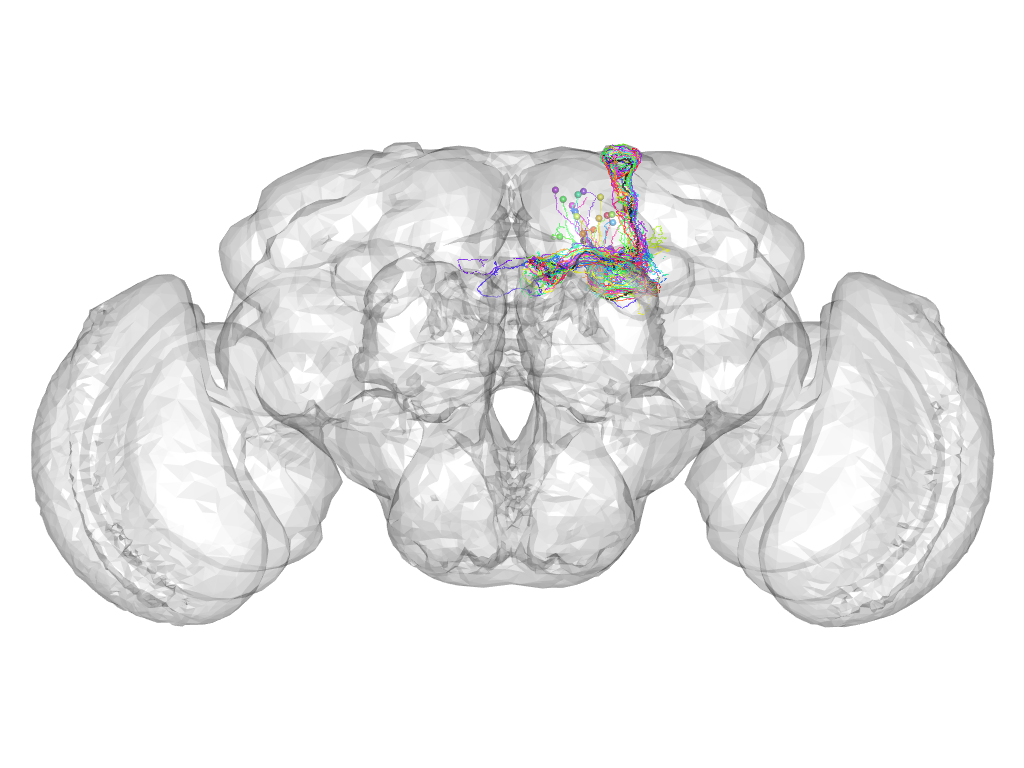
Cluster 407
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-600073), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 533 | Cha-F-900008 | XVI | 0.60 |
| 125 | fru-M-000004 | XVI | 0.58 |
| 522 | Cha-F-300056 | XVI | 0.57 |
| 26 | fru-M-200296 | XVI | 0.56 |
| 419 | fru-F-200021 | XVI | 0.54 |
| 53 | fru-M-000099 | XVI | 0.51 |
Cluster composition
This cluster has 24 neurons. The exemplar of this cluster (fru-F-600073) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-600073 | 6302 | FruMARCM-F001424_seg001 | fru-Gal4 | F | 7858.57 | 1.00 |
| fru-M-300134 | 1047 | FruMARCM-M001226_seg003 | fru-Gal4 | M | 4865.83 | 0.63 |
| fru-M-600036 | 1410 | FruMARCM-M000907_seg002 | fru-Gal4 | M | 4992.57 | 0.62 |
| fru-F-500201 | 4532 | FruMARCM-F001641_seg002 | fru-Gal4 | F | 4613.10 | 0.58 |
| fru-F-500243 | 4734 | FruMARCM-F002059_seg001 | fru-Gal4 | F | 5185.79 | 0.65 |
| VGlut-F-500755 | 6816 | DvGlutMARCM-F004002_seg001 | VGlut-Gal4 | F | 4287.39 | 0.51 |
| VGlut-F-700480 | 6985 | DvGlutMARCM-F004131_seg001 | VGlut-Gal4 | F | 4452.76 | 0.56 |
| fru-F-600048 | 7900 | FruMARCM-F000845_seg001 | fru-Gal4 | F | 4663.24 | 0.59 |
| Cha-F-300109 | 8364 | ChaMARCM-F000308_seg001 | Cha-Gal4 | F | 4709.29 | 0.56 |
| Gad1-F-700006 | 8402 | GadMARCM-F000130_seg002 | Gad1-Gal4 | F | 4540.86 | 0.52 |
| VGlut-F-600517 | 8549 | DvGlutMARCM-F003119_seg002 | VGlut-Gal4 | F | 4682.32 | 0.57 |
| VGlut-F-400738 | 8823 | DvGlutMARCM-F003626_seg001 | VGlut-Gal4 | F | 4876.06 | 0.58 |
| Gad1-F-000037 | 9510 | GadMARCM-F000401_seg002 | Gad1-Gal4 | F | 5089.12 | 0.64 |
| Gad1-F-500032 | 10516 | GadMARCM-F000159_seg001 | Gad1-Gal4 | F | 4559.84 | 0.58 |
| Gad1-F-300065 | 11235 | GadMARCM-F000217_seg001 | Gad1-Gal4 | F | 4809.72 | 0.61 |
| VGlut-F-800122 | 11562 | DvGlutMARCM-F003036_seg002 | VGlut-Gal4 | F | 4466.31 | 0.55 |
| Cha-F-700000 | 11598 | ChaMARCM-F000027_seg001 | Cha-Gal4 | F | 5275.83 | 0.66 |
| VGlut-F-100244 | 11652 | DvGlutMARCM-F002226_seg001 | VGlut-Gal4 | F | 4621.38 | 0.51 |
| VGlut-F-000420 | 12850 | DvGlutMARCM-F1895_seg1 | VGlut-Gal4 | F | 4844.75 | 0.55 |
| VGlut-F-400471 | 12929 | DvGlutMARCM-F1967_seg2 | VGlut-Gal4 | F | 4963.37 | 0.61 |
| VGlut-F-100163 | 15216 | DvGlutMARCM-F1542_seg1 | VGlut-Gal4 | F | 5149.56 | 0.63 |
| VGlut-F-500192 | 15227 | DvGlutMARCM-F1550_seg1 | VGlut-Gal4 | F | 5499.31 | 0.68 |
| VGlut-F-500258 | 15425 | DvGlutMARCM-F1781_seg2 | VGlut-Gal4 | F | 4640.83 | 0.59 |
| VGlut-F-200098 | 16136 | DvGlutMARCM-F518_seg1 | VGlut-Gal4 | F | 4148.77 | 0.54 |