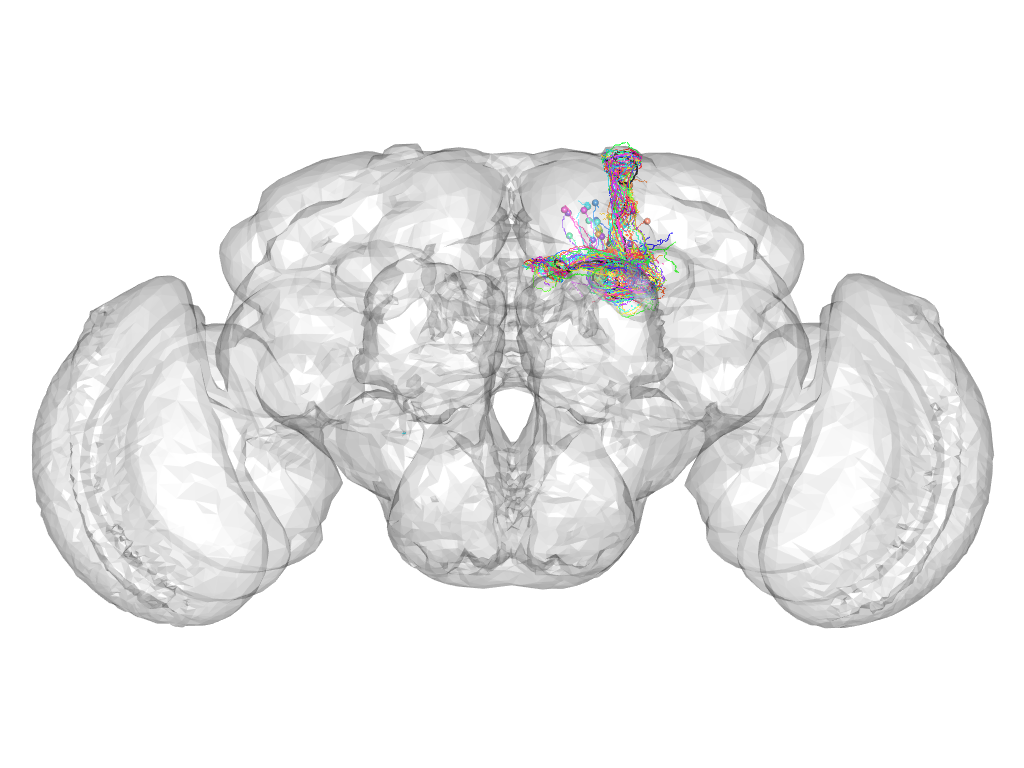
Cluster 533
Part of supercluster XVI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Cha-F-900008), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 407 | fru-F-600073 | XVI | 0.61 |
| 26 | fru-M-200296 | XVI | 0.59 |
| 522 | Cha-F-300056 | XVI | 0.58 |
| 125 | fru-M-000004 | XVI | 0.54 |
| 419 | fru-F-200021 | XVI | 0.53 |
| 53 | fru-M-000099 | XVI | 0.52 |
Cluster composition
This cluster has 32 neurons. The exemplar of this cluster (Cha-F-900008) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Cha-F-900008 | 8332 | ChaMARCM-F000287_seg002 | Cha-Gal4 | F | 6936.04 | 1.00 |
| fru-M-600050 | 1171 | FruMARCM-M001355_seg002 | fru-Gal4 | M | 4616.76 | 0.66 |
| 5-HT1B-M-700002 | 2502 | 5HT1bMARCM-M000048_seg001 | 5-HT1B-Gal4 | M | 3601.33 | 0.54 |
| Gad1-F-000095 | 4107 | GadMARCM-F000693_seg001 | Gad1-Gal4 | F | 3725.34 | 0.47 |
| fru-F-500218 | 4659 | FruMARCM-F001821_seg003 | fru-Gal4 | F | 4469.27 | 0.63 |
| Cha-F-400181 | 5467 | ChaMARCM-F000799_seg003 | Cha-Gal4 | F | 3819.83 | 0.54 |
| VGlut-F-500800 | 6522 | DvGlutMARCM-F004294_seg001 | VGlut-Gal4 | F | 4069.34 | 0.57 |
| Cha-F-400124 | 7118 | ChaMARCM-F000591_seg001 | Cha-Gal4 | F | 4408.71 | 0.64 |
| Cha-F-300165 | 7145 | ChaMARCM-F000609_seg002 | Cha-Gal4 | F | 4105.55 | 0.60 |
| Cha-F-500133 | 7198 | ChaMARCM-F000644_seg001 | Cha-Gal4 | F | 2448.38 | 0.43 |
| Cha-F-500102 | 7236 | ChaMARCM-F000410_seg005 | Cha-Gal4 | F | 4340.05 | 0.61 |
| Cha-F-100043 | 7614 | ChaMARCM-F000458_seg001 | Cha-Gal4 | F | 3402.77 | 0.50 |
| VGlut-F-600585 | 9263 | DvGlutMARCM-F003320_seg001 | VGlut-Gal4 | F | 4353.92 | 0.61 |
| Gad1-F-300103 | 9529 | GadMARCM-F000412_seg001 | Gad1-Gal4 | F | 4197.89 | 0.62 |
| Gad1-F-800043 | 9549 | GadMARCM-F000429_seg001 | Gad1-Gal4 | F | 4446.03 | 0.64 |
| fru-F-700030 | 9782 | FruMARCM-F000298_seg002 | fru-Gal4 | F | 4781.62 | 0.64 |
| fru-F-500063 | 9983 | FruMARCM-F000571_seg001 | fru-Gal4 | F | 4255.65 | 0.60 |
| fru-F-500076 | 10002 | FruMARCM-F000584_seg003 | fru-Gal4 | F | 2964.34 | 0.46 |
| fru-F-600042 | 10010 | FruMARCM-F000589_seg002 | fru-Gal4 | F | 4280.44 | 0.62 |
| Gad1-F-700002 | 10609 | GadMARCM-F000117_seg001 | Gad1-Gal4 | F | 4300.11 | 0.61 |
| Gad1-F-200041 | 10679 | GadMARCM-F000184_seg001 | Gad1-Gal4 | F | 4278.38 | 0.61 |
| VGlut-F-600439 | 10965 | DvGlutMARCM-F002858_seg002 | VGlut-Gal4 | F | 4090.98 | 0.60 |
| VGlut-F-200445 | 11085 | DvGlutMARCM-F002958_seg002 | VGlut-Gal4 | F | 1665.50 | 0.38 |
| Gad1-F-900026 | 11298 | GadMARCM-F000272_seg001 | Gad1-Gal4 | F | 3380.72 | 0.53 |
| Gad1-F-200079 | 11325 | GadMARCM-F000296_seg001 | Gad1-Gal4 | F | 3674.22 | 0.51 |
| Gad1-F-600049 | 11424 | GadMARCM-F000363_seg001 | Gad1-Gal4 | F | 4570.29 | 0.66 |
| Gad1-F-200002 | 12613 | GadMARCM-F000017_seg001 | Gad1-Gal4 | F | 3964.21 | 0.55 |
| VGlut-F-500309 | 12889 | DvGlutMARCM-F1932_seg1 | VGlut-Gal4 | F | 4493.83 | 0.62 |
| VGlut-F-400286 | 14332 | DvGlutMARCM-F1014_seg1 | VGlut-Gal4 | F | 4453.54 | 0.63 |
| VGlut-F-100074 | 14730 | DvGlutMARCM-F990_seg1 | VGlut-Gal4 | F | 4291.75 | 0.61 |
| VGlut-F-000302 | 14744 | DvGlutMARCM-F1095_seg2 | VGlut-Gal4 | F | 4529.21 | 0.58 |
| VGlut-F-300236 | 14971 | DvGlutMARCM-F1309_seg1 | VGlut-Gal4 | F | 3957.99 | 0.55 |