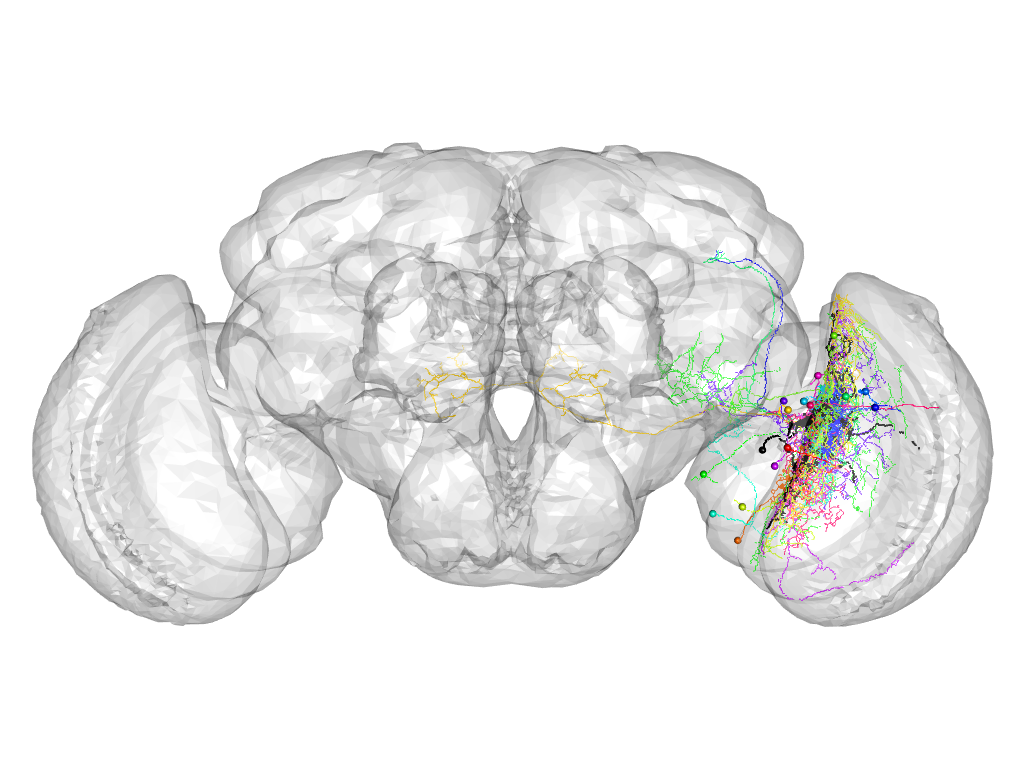
Cluster 1026
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-400078), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 1020 | VGlut-F-100029 | III | 0.74 |
| 1040 | VGlut-F-200092 | III | 0.73 |
| 173 | Trh-M-000051 | III | 0.69 |
| 16 | fru-M-300276 | III | 0.69 |
| 661 | Tdc2-F-000027 | VII | 0.68 |
| 874 | Trh-F-100011 | III | 0.65 |
| 1006 | VGlut-F-500043 | III | 0.61 |
| 919 | VGlut-F-000205 | III | 0.59 |
| 952 | VGlut-F-600052 | III | 0.59 |
| 759 | VGlut-F-600257 | III | 0.59 |
| 175 | Trh-M-700055 | III | 0.58 |
| 21 | fru-M-300286 | III | 0.55 |
| 202 | Trh-M-100011 | III | 0.53 |
| 849 | TH-F-300018 | I | 0.51 |
| 468 | VGlut-F-600670 | III | 0.51 |
Cluster composition
This cluster has 16 neurons. The exemplar of this cluster (VGlut-F-400078) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-400078 | 15981 | DvGlutMARCM-F372_seg1 | VGlut-Gal4 | F | 29065.31 | 1.00 |
| Gad1-F-800066 | 3956 | GadMARCM-F000583_seg002 | Gad1-Gal4 | F | 10248.34 | 0.52 |
| Gad1-F-300139 | 4018 | GadMARCM-F000629_seg001 | Gad1-Gal4 | F | 6873.66 | 0.38 |
| Cha-F-000186 | 4965 | ChaMARCM-F001063_seg001 | Cha-Gal4 | F | 20773.06 | 0.42 |
| Cha-F-500171 | 5063 | ChaMARCM-F001136_seg002 | Cha-Gal4 | F | 17485.12 | 0.56 |
| Cha-F-000267 | 5201 | ChaMARCM-F001235_seg001 | Cha-Gal4 | F | 12202.29 | 0.43 |
| Cha-F-000140 | 5398 | ChaMARCM-F000755_seg002 | Cha-Gal4 | F | 16070.73 | 0.23 |
| Cha-F-400194 | 5563 | ChaMARCM-F000863_seg001 | Cha-Gal4 | F | 10660.21 | 0.35 |
| Cha-F-200044 | 8223 | ChaMARCM-F000224_seg001 | Cha-Gal4 | F | 8798.93 | 0.32 |
| VGlut-F-700392 | 8734 | DvGlutMARCM-F003564_seg002 | VGlut-Gal4 | F | 10303.38 | 0.55 |
| VGlut-F-200484 | 8997 | DvGlutMARCM-F003544_seg001 | VGlut-Gal4 | F | 6468.24 | 0.42 |
| Gad1-F-400023 | 11243 | GadMARCM-F000224_seg001 | Gad1-Gal4 | F | 9323.56 | 0.41 |
| VGlut-F-400574 | 12259 | DvGlutMARCM-F002719_seg001 | VGlut-Gal4 | F | 19230.89 | 0.52 |
| VGlut-F-300389 | 13118 | DvGlutMARCM-F2144_seg1 | VGlut-Gal4 | F | 20066.63 | 0.55 |
| VGlut-F-000076 | 16013 | DvGlutMARCM-F401_seg1 | VGlut-Gal4 | F | 17609.52 | 0.65 |
| VGlut-F-100046 | 16137 | DvGlutMARCM-F520_seg1 | VGlut-Gal4 | F | 11958.97 | 0.32 |