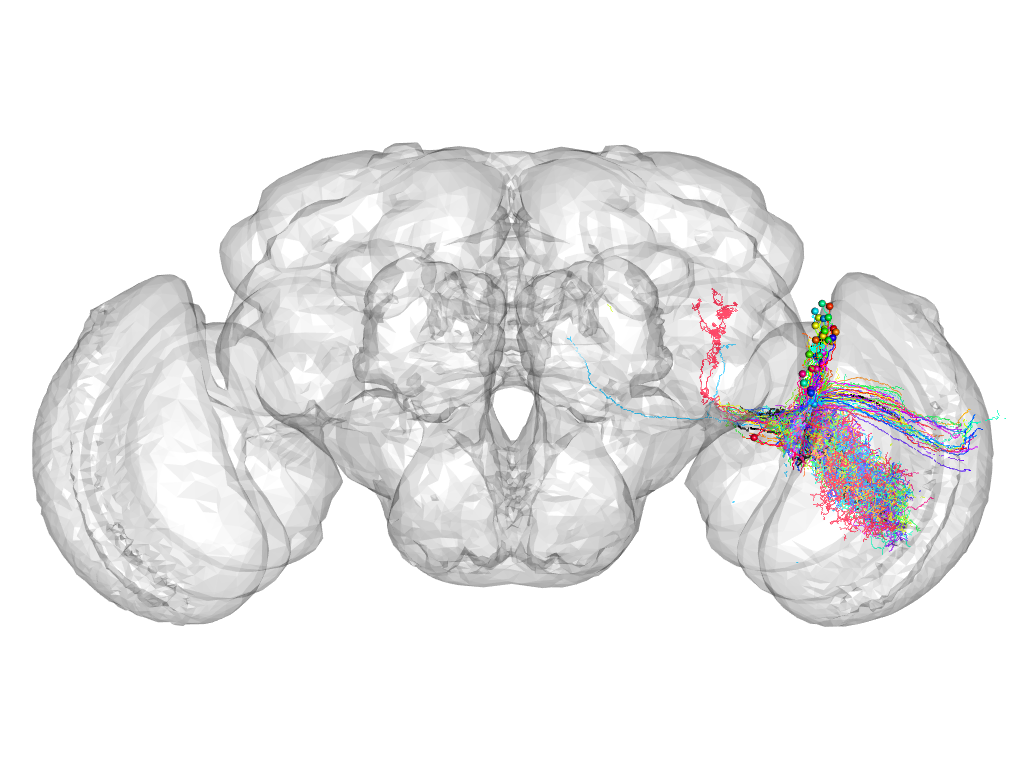
Cluster 202
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-100011), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 173 | Trh-M-000051 | III | 0.66 |
| 874 | Trh-F-100011 | III | 0.64 |
| 661 | Tdc2-F-000027 | VII | 0.62 |
| 4 | fru-M-500226 | III | 0.62 |
| 150 | Trh-M-000085 | III | 0.60 |
| 21 | fru-M-300286 | III | 0.59 |
| 1006 | VGlut-F-500043 | III | 0.55 |
| 76 | fru-M-300240 | III | 0.54 |
| 759 | VGlut-F-600257 | III | 0.54 |
| 952 | VGlut-F-600052 | III | 0.52 |
Cluster composition
This cluster has 36 neurons. The exemplar of this cluster (Trh-M-100011) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-100011 | 3392 | TPHMARCM-075M_seg1 | Trh-Gal4 | M | 15295.74 | 1.00 |
| fru-M-800093 | 83 | FruMARCM-M002282_seg001 | fru-Gal4 | M | 10552.11 | 0.66 |
| fru-M-600100 | 87 | FruMARCM-M002286_seg001 | fru-Gal4 | M | 8682.57 | 0.63 |
| fru-M-900051 | 105 | FruMARCM-M002304_seg001 | fru-Gal4 | M | 10649.18 | 0.69 |
| fru-M-200277 | 113 | FruMARCM-M002311_seg001 | fru-Gal4 | M | 9771.57 | 0.65 |
| fru-M-200301 | 269 | FruMARCM-M002495_seg001 | fru-Gal4 | M | 10769.37 | 0.68 |
| fru-M-700107 | 670 | FruMARCM-M001877_seg001 | fru-Gal4 | M | 10460.98 | 0.67 |
| fru-M-400055 | 1686 | FruMARCM-M000840_seg001 | fru-Gal4 | M | 9651.30 | 0.65 |
| fru-M-100040 | 1879 | FruMARCM-M000420_seg001 | fru-Gal4 | M | 11009.40 | 0.66 |
| fru-M-300056 | 1901 | FruMARCM-M000437_seg001 | fru-Gal4 | M | 10323.45 | 0.66 |
| Trh-M-900041 | 2051 | TPHMARCM-M001556_seg001 | Trh-Gal4 | M | 10839.50 | 0.70 |
| Trh-M-000076 | 2246 | TPHMARCM-M001733_seg001 | Trh-Gal4 | M | 11084.32 | 0.68 |
| fru-M-200001 | 2320 | FruMARCM-M000006_seg001 | fru-Gal4 | M | 8369.29 | 0.54 |
| fru-M-900010 | 2385 | FruMARCM-M000112_seg001 | fru-Gal4 | M | 11091.95 | 0.71 |
| fru-M-600006 | 2414 | FruMARCM-M000135_seg001 | fru-Gal4 | M | 10719.24 | 0.65 |
| Trh-M-500071 | 3005 | TPHMARCM-949M_seg1 | Trh-Gal4 | M | 10230.74 | 0.65 |
| Trh-M-700000 | 3432 | TPHMARCM-175M_seg1 | Trh-Gal4 | M | 10942.35 | 0.66 |
| Trh-M-500010 | 3447 | TPHMARCM-196M_seg1 | Trh-Gal4 | M | 10411.37 | 0.62 |
| Trh-M-100031 | 3471 | TPHMARCM-341M_seg1 | Trh-Gal4 | M | 11101.60 | 0.70 |
| Trh-M-700001 | 3591 | TPHMARCM-535M_seg1 | Trh-Gal4 | M | 11049.81 | 0.67 |
| Cha-F-300026 | 3700 | ChaMARCM-F000112_seg001 | Cha-Gal4 | F | 8318.25 | 0.46 |
| fru-F-500271 | 3739 | FruMARCM-F002332_seg001 | fru-Gal4 | F | 9657.29 | 0.61 |
| fru-F-100089 | 3804 | FruMARCM-F002559_seg001 | fru-Gal4 | F | 11030.70 | 0.69 |
| fru-F-800055 | 4642 | FruMARCM-F001809_seg001 | fru-Gal4 | F | 10949.91 | 0.68 |
| fru-F-400221 | 4802 | FruMARCM-F002136_seg001 | fru-Gal4 | F | 11353.88 | 0.69 |
| Tdc2-F-100000 | 8441 | dTdc2MARCM-F002_seg1 | Tdc2-Gal4 | F | 10680.44 | 0.70 |
| fru-F-000008 | 11542 | FruMARCM-F000199_seg001 | fru-Gal4 | F | 9683.61 | 0.59 |
| Trh-F-100004 | 13787 | TPHMARCM-005F_seg1 | Trh-Gal4 | F | 10140.11 | 0.63 |
| Trh-F-200002 | 13796 | TPHMARCM-014F_seg1 | Trh-Gal4 | F | 10659.31 | 0.66 |
| Trh-F-100010 | 13797 | TPHMARCM-015F_seg1 | Trh-Gal4 | F | 10615.04 | 0.67 |
| Trh-F-000003 | 13800 | TPHMARCM-018F_seg1 | Trh-Gal4 | F | 10829.57 | 0.67 |
| Trh-F-300001 | 13822 | TPHMARCM-043F_seg1 | Trh-Gal4 | F | 8811.64 | 0.59 |
| Trh-F-900005 | 13924 | TPHMARCM-215F_seg1 | Trh-Gal4 | F | 9451.54 | 0.68 |
| Trh-F-900009 | 13928 | TPHMARCM-218F_seg1 | Trh-Gal4 | F | 10748.74 | 0.63 |
| Trh-F-100054 | 14130 | TPHMARCM-518F_seg2 | Trh-Gal4 | F | 10650.26 | 0.66 |
| VGlut-F-200326 | 15384 | DvGlutMARCM-F1741_seg1 | VGlut-Gal4 | F | 8721.48 | 0.36 |