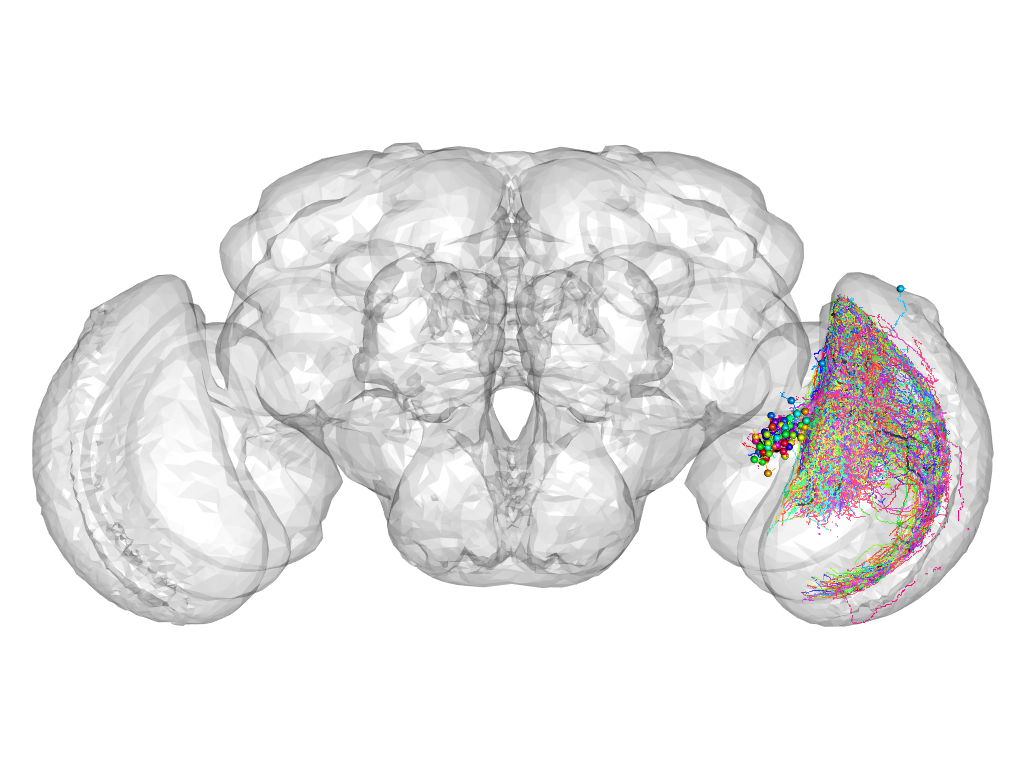
Cluster 16
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-300276), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 661 | Tdc2-F-000027 | VII | 0.74 |
| 173 | Trh-M-000051 | III | 0.68 |
| 952 | VGlut-F-600052 | III | 0.66 |
| 759 | VGlut-F-600257 | III | 0.65 |
| 1020 | VGlut-F-100029 | III | 0.64 |
| 1006 | VGlut-F-500043 | III | 0.63 |
| 874 | Trh-F-100011 | III | 0.61 |
| 468 | VGlut-F-600670 | III | 0.58 |
| 1040 | VGlut-F-200092 | III | 0.56 |
| 175 | Trh-M-700055 | III | 0.55 |
Cluster composition
This cluster has 79 neurons. The exemplar of this cluster (fru-M-300276) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-300276 | 202 | FruMARCM-M002417_seg001 | fru-Gal4 | M | 29999.23 | 1.00 |
| fru-M-200264 | 53 | FruMARCM-M002249_seg001 | fru-Gal4 | M | 22171.32 | 0.73 |
| fru-M-200272 | 93 | FruMARCM-M002292_seg001 | fru-Gal4 | M | 21486.31 | 0.69 |
| fru-M-300261 | 137 | FruMARCM-M002349_seg001 | fru-Gal4 | M | 20523.55 | 0.68 |
| fru-M-700159 | 150 | FruMARCM-M002376_seg001 | fru-Gal4 | M | 21303.44 | 0.69 |
| fru-M-900060 | 158 | FruMARCM-M002380_seg001 | fru-Gal4 | M | 20064.56 | 0.68 |
| fru-M-700171 | 201 | FruMARCM-M002416_seg001 | fru-Gal4 | M | 22242.67 | 0.72 |
| fru-M-300277 | 204 | FruMARCM-M002419_seg001 | fru-Gal4 | M | 20688.23 | 0.68 |
| fru-M-700090 | 468 | FruMARCM-M001573_seg001 | fru-Gal4 | M | 19920.75 | 0.65 |
| fru-M-400161 | 512 | FruMARCM-M001614_seg001 | fru-Gal4 | M | 21033.07 | 0.70 |
| fru-M-000105 | 582 | FruMARCM-M001773_seg001 | fru-Gal4 | M | 21158.28 | 0.71 |
| fru-M-800067 | 585 | FruMARCM-M001778_seg001 | fru-Gal4 | M | 20800.49 | 0.69 |
| fru-M-700099 | 619 | FruMARCM-M001806_seg001 | fru-Gal4 | M | 18642.13 | 0.67 |
| fru-M-700113 | 706 | FruMARCM-M001909_seg001 | fru-Gal4 | M | 21859.36 | 0.73 |
| fru-M-700114 | 707 | FruMARCM-M001910_seg001 | fru-Gal4 | M | 21939.85 | 0.73 |
| fru-M-600038 | 982 | FruMARCM-M001161_seg001 | fru-Gal4 | M | 21362.26 | 0.70 |
| fru-M-200148 | 991 | FruMARCM-M001173_seg001 | fru-Gal4 | M | 20210.58 | 0.68 |
| fru-M-700082 | 1257 | FruMARCM-M001450_seg001 | fru-Gal4 | M | 20171.34 | 0.67 |
| fru-M-700060 | 1380 | FruMARCM-M000886_seg001 | fru-Gal4 | M | 21815.27 | 0.67 |
| fru-M-600030 | 1384 | FruMARCM-M000890_seg001 | fru-Gal4 | M | 21807.29 | 0.72 |
| fru-M-500081 | 1395 | FruMARCM-M000896_seg002 | fru-Gal4 | M | 20683.45 | 0.68 |
| fru-M-100093 | 1531 | FruMARCM-M001070_seg001 | fru-Gal4 | M | 21144.26 | 0.69 |
| fru-M-200135 | 1549 | FruMARCM-M001082_seg001 | fru-Gal4 | M | 20267.38 | 0.68 |
| fru-M-800022 | 1581 | FruMARCM-M000651_seg001 | fru-Gal4 | M | 20740.28 | 0.70 |
| fru-M-200081 | 1601 | FruMARCM-M000696_seg001 | fru-Gal4 | M | 19493.60 | 0.63 |
| fru-M-200083 | 1616 | FruMARCM-M000729_seg001 | fru-Gal4 | M | 19129.23 | 0.64 |
| fru-M-100065 | 1646 | FruMARCM-M000786_seg002 | fru-Gal4 | M | 19861.95 | 0.65 |
| fru-M-100066 | 1647 | FruMARCM-M000787_seg001 | fru-Gal4 | M | 20727.17 | 0.67 |
| fru-M-200049 | 1793 | FruMARCM-M000321_seg001 | fru-Gal4 | M | 18077.76 | 0.66 |
| fru-M-700003 | 2020 | FruMARCM-M000147_seg005 | fru-Gal4 | M | 17989.31 | 0.63 |
| fru-M-900009 | 2384 | FruMARCM-M000111_seg001 | fru-Gal4 | M | 21759.90 | 0.70 |
| fru-F-000093 | 4619 | FruMARCM-F001745_seg001 | fru-Gal4 | F | 20124.31 | 0.69 |
| fru-F-700159 | 4660 | FruMARCM-F001822_seg001 | fru-Gal4 | F | 21891.07 | 0.71 |
| fru-F-300114 | 4764 | FruMARCM-F002083_seg001 | fru-Gal4 | F | 20796.17 | 0.71 |
| VGlut-F-300658 | 5928 | DvGlutMARCM-F004588_seg001 | VGlut-Gal4 | F | 19774.30 | 0.65 |
| fru-F-200078 | 6219 | FruMARCM-F001142_seg001 | fru-Gal4 | F | 21058.03 | 0.68 |
| fru-F-000044 | 6263 | FruMARCM-F001299_seg001 | fru-Gal4 | F | 20374.21 | 0.71 |
| VGlut-F-300600 | 6413 | DvGlutMARCM-F004205_seg001 | VGlut-Gal4 | F | 19894.48 | 0.66 |
| VGlut-F-300568 | 6784 | DvGlutMARCM-F003979_seg001 | VGlut-Gal4 | F | 19749.69 | 0.65 |
| VGlut-F-300579 | 6898 | DvGlutMARCM-F004054_seg001 | VGlut-Gal4 | F | 21312.70 | 0.71 |
| VGlut-F-000610 | 7439 | DvGlutMARCM-F003892_seg001 | VGlut-Gal4 | F | 19552.87 | 0.68 |
| fru-F-900014 | 7878 | FruMARCM-F000801_seg001 | fru-Gal4 | F | 21273.44 | 0.68 |
| VGlut-F-300531 | 7906 | DvGlutMARCM-F003752_seg001 | VGlut-Gal4 | F | 17785.73 | 0.61 |
| Cha-F-700081 | 8253 | ChaMARCM-F000241_seg002 | Cha-Gal4 | F | -1513.10 | 0.29 |
| VGlut-F-300514 | 8601 | DvGlutMARCM-F003664_seg001 | VGlut-Gal4 | F | 18723.49 | 0.68 |
| VGlut-F-300516 | 8606 | DvGlutMARCM-F003668_seg001 | VGlut-Gal4 | F | 17457.71 | 0.61 |
| VGlut-F-900087 | 8901 | DvGlutMARCM-F003477_seg001 | VGlut-Gal4 | F | 16540.57 | 0.62 |
| VGlut-F-600654 | 8969 | DvGlutMARCM-F003528_seg001 | VGlut-Gal4 | F | 21276.83 | 0.71 |
| VGlut-F-100279 | 9216 | DvGlutMARCM-F003282_seg001 | VGlut-Gal4 | F | 20539.86 | 0.67 |
| VGlut-F-100286 | 9267 | DvGlutMARCM-F003325_seg001 | VGlut-Gal4 | F | 19950.63 | 0.66 |
| VGlut-F-800138 | 9302 | DvGlutMARCM-F003149_seg001 | VGlut-Gal4 | F | 20812.63 | 0.68 |
| fru-F-800009 | 9746 | FruMARCM-F000260_seg001 | fru-Gal4 | F | 20110.92 | 0.67 |
| fru-F-500005 | 9753 | FruMARCM-F000267_seg001 | fru-Gal4 | F | 21870.97 | 0.71 |
| VGlut-F-300328 | 10241 | DvGlutMARCM-F1735_seg1 | VGlut-Gal4 | F | 21730.78 | 0.69 |
| VGlut-F-400608 | 10887 | DvGlutMARCM-F002793_seg001 | VGlut-Gal4 | F | 20024.21 | 0.67 |
| VGlut-F-700228 | 11053 | DvGlutMARCM-F002932_seg001 | VGlut-Gal4 | F | 20468.72 | 0.69 |
| VGlut-F-400638 | 11664 | DvGlutMARCM-F002243_seg001 | VGlut-Gal4 | F | 20519.61 | 0.69 |
| VGlut-F-400511 | 11700 | DvGlutMARCM-F002273_seg001 | VGlut-Gal4 | F | 18123.78 | 0.65 |
| VGlut-F-000467 | 12080 | DvGlutMARCM-F002589_seg001 | VGlut-Gal4 | F | 18201.81 | 0.66 |
| VGlut-F-300390 | 13119 | DvGlutMARCM-F2145_seg1 | VGlut-Gal4 | F | 21086.12 | 0.69 |
| VGlut-F-100231 | 13128 | DvGlutMARCM-F2155_seg1 | VGlut-Gal4 | F | 21732.00 | 0.72 |
| VGlut-F-300394 | 13151 | DvGlutMARCM-F2184_seg1 | VGlut-Gal4 | F | 21138.53 | 0.70 |
| VGlut-F-300148 | 14440 | DvGlutMARCM-F731_seg1 | VGlut-Gal4 | F | 19196.04 | 0.67 |
| VGlut-F-200185 | 14683 | DvGlutMARCM-F943_seg1 | VGlut-Gal4 | F | 20332.72 | 0.65 |
| VGlut-F-500153 | 14742 | TMARCM-F1371_seg1 | VGlut-Gal4 | F | 20004.68 | 0.67 |
| VGlut-F-000315 | 14832 | DvGlutMARCM-F1175_seg1 | VGlut-Gal4 | F | 22066.92 | 0.70 |
| VGlut-F-500152 | 14949 | DvGlutMARCM-F1287_seg1 | VGlut-Gal4 | F | 16047.32 | 0.61 |
| VGlut-F-900025 | 15024 | DvGlutMARCM-F1363_seg1 | VGlut-Gal4 | F | 21951.07 | 0.70 |
| VGlut-F-200141 | 15553 | DvGlutMARCM-F689-X2_seg1 | VGlut-Gal4 | F | 18131.50 | 0.64 |
| VGlut-F-300007 | 15678 | DvGlutMARCM-F093_seg1 | VGlut-Gal4 | F | 19200.82 | 0.65 |
| VGlut-F-200055 | 15690 | DvGlutMARCM-F105_seg1 | VGlut-Gal4 | F | 19543.94 | 0.63 |
| VGlut-F-000020 | 15763 | DvGlutMARCM-F170_seg1 | VGlut-Gal4 | F | 21257.06 | 0.70 |
| VGlut-F-000021 | 15764 | DvGlutMARCM-F171_seg1 | VGlut-Gal4 | F | 21024.52 | 0.70 |
| VGlut-F-200074 | 15809 | DvGlutMARCM-F217_seg1 | VGlut-Gal4 | F | 20908.48 | 0.62 |
| VGlut-F-000037 | 15844 | DvGlutMARCM-F251_seg1 | VGlut-Gal4 | F | 20863.22 | 0.66 |
| VGlut-F-000045 | 15866 | DvGlutMARCM-F269_seg1 | VGlut-Gal4 | F | 19413.38 | 0.62 |
| VGlut-F-000102 | 16092 | DvGlutMARCM-F476_seg1 | VGlut-Gal4 | F | 21049.55 | 0.68 |
| VGlut-F-000112 | 16103 | DvGlutMARCM-F486_seg1 | VGlut-Gal4 | F | 18130.31 | 0.59 |
| VGlut-F-300083 | 16157 | DvGlutMARCM-F539_seg1 | VGlut-Gal4 | F | 21492.81 | 0.69 |