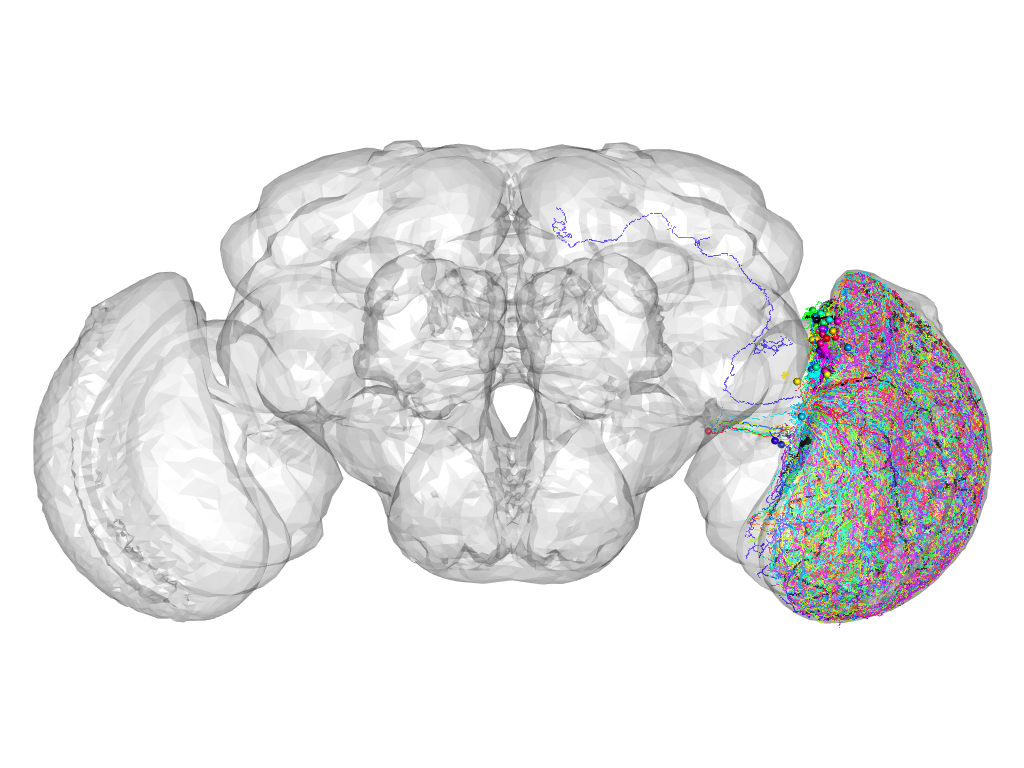
Cluster 175
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-700055), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 468 | VGlut-F-600670 | III | 0.54 |
| 209 | Trh-M-600023 | III | 0.50 |
| 759 | VGlut-F-600257 | III | 0.46 |
| 660 | Tdc2-F-200049 | I | 0.41 |
| 661 | Tdc2-F-000027 | VII | 0.37 |
Cluster composition
This cluster has 61 neurons. The exemplar of this cluster (Trh-M-700055) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-700055 | 2804 | TPHMARCM-1033M_seg1 | Trh-Gal4 | M | 363828.94 | 1.00 |
| Trh-M-000000 | 954 | TPHMARCM-062M_seg1 | Trh-Gal4 | M | 222551.32 | 0.63 |
| Trh-M-000133 | 1725 | TPHMARCM-M001801_seg001 | Trh-Gal4 | M | 231261.69 | 0.65 |
| Trh-M-000151 | 1742 | TPHMARCM-M001819_seg001 | Trh-Gal4 | M | 238142.06 | 0.65 |
| Trh-M-500076 | 1991 | TPHMARCM-0982M_seg001 | Trh-Gal4 | M | 212642.60 | 0.62 |
| Trh-M-600106 | 2032 | TPHMARCM-M001697_seg001 | Trh-Gal4 | M | 205600.91 | 0.60 |
| Trh-M-900043 | 2053 | TPHMARCM-M001558_seg001 | Trh-Gal4 | M | 216779.03 | 0.64 |
| Trh-M-600097 | 2197 | TPHMARCM-M001687_seg001 | Trh-Gal4 | M | 227526.26 | 0.64 |
| Trh-M-000089 | 2263 | TPHMARCM-M001750_seg001 | Trh-Gal4 | M | 229027.40 | 0.65 |
| Trh-M-000090 | 2264 | TPHMARCM-M001751_seg001 | Trh-Gal4 | M | 235421.66 | 0.67 |
| Trh-M-000097 | 2285 | TPHMARCM-M001766_seg001 | Trh-Gal4 | M | 252900.71 | 0.72 |
| Trh-M-000098 | 2286 | TPHMARCM-M001767_seg001 | Trh-Gal4 | M | 242843.95 | 0.69 |
| Trh-M-000102 | 2289 | TPHMARCM-M001771_seg001 | Trh-Gal4 | M | 212371.52 | 0.60 |
| Trh-M-000110 | 2296 | TPHMARCM-M001778_seg001 | Trh-Gal4 | M | 214907.34 | 0.63 |
| Trh-M-000126 | 2312 | TPHMARCM-M001794_seg001 | Trh-Gal4 | M | 206467.69 | 0.59 |
| Trh-M-000128 | 2314 | TPHMARCM-M001796_seg001 | Trh-Gal4 | M | 209746.48 | 0.61 |
| Trh-M-700076 | 2607 | TPHMARCM-1289M_seg1 | Trh-Gal4 | M | 231005.48 | 0.66 |
| Trh-M-700079 | 2615 | TPHMARCM-1325M_seg1 | Trh-Gal4 | M | 225692.43 | 0.64 |
| Trh-M-000029 | 2655 | TPHMARCM-M001423_seg001 | Trh-Gal4 | M | 230094.58 | 0.66 |
| Trh-M-000036 | 2674 | TPHMARCM-M001440_seg001 | Trh-Gal4 | M | 232681.42 | 0.66 |
| Trh-M-000038 | 2676 | TPHMARCM-M001442_seg001 | Trh-Gal4 | M | 233299.09 | 0.66 |
| Trh-M-000045 | 2690 | TPHMARCM-M001457_seg001 | Trh-Gal4 | M | 217695.17 | 0.65 |
| Trh-M-000048 | 2693 | TPHMARCM-M001460_seg1 | Trh-Gal4 | M | 240178.28 | 0.67 |
| Trh-M-800021 | 2751 | TPHMARCM-M001517_seg001 | Trh-Gal4 | M | 230711.81 | 0.66 |
| Trh-M-700044 | 2792 | TPHMARCM-1020M_seg1 | Trh-Gal4 | M | 233785.59 | 0.66 |
| Trh-M-700050 | 2796 | TPHMARCM-1024M_seg1 | Trh-Gal4 | M | 235753.23 | 0.67 |
| Trh-M-700056 | 2805 | TPHMARCM-1034M_seg1 | Trh-Gal4 | M | 222851.58 | 0.63 |
| Trh-M-600060 | 2812 | TPHMARCM-1041M_seg1 | Trh-Gal4 | M | 199815.64 | 0.58 |
| Trh-M-500087 | 2858 | TPHMARCM-1087M_seg1 | Trh-Gal4 | M | 221651.10 | 0.63 |
| Trh-M-200045 | 2986 | TPHMARCM-923M_seg1 | Trh-Gal4 | M | 228619.70 | 0.66 |
| Trh-M-600025 | 3385 | TPHMARCM-585M_seg1 | Trh-Gal4 | M | 228734.96 | 0.64 |
| Trh-M-100013 | 3394 | TPHMARCM-078M_seg1 | Trh-Gal4 | M | 224899.69 | 0.63 |
| Trh-M-100015 | 3396 | TPHMARCM-081M_seg1 | Trh-Gal4 | M | 165742.89 | 0.54 |
| Trh-M-000005 | 3400 | TPHMARCM-086M_seg1 | Trh-Gal4 | M | 207107.68 | 0.59 |
| Trh-M-100017 | 3401 | TPHMARCM-087M_seg1 | Trh-Gal4 | M | 226504.47 | 0.62 |
| Trh-M-900002 | 3452 | TPHMARCM-208M_seg1 | Trh-Gal4 | M | 228859.04 | 0.65 |
| Trh-M-900006 | 3587 | TPHMARCM-532M_seg1 | Trh-Gal4 | M | 247341.04 | 0.69 |
| Trh-M-600016 | 3600 | TPHMARCM-543M_seg1 | Trh-Gal4 | M | 230483.35 | 0.67 |
| Trh-M-600022 | 3612 | TPHMARCM-572M_seg1 | Trh-Gal4 | M | 219659.39 | 0.62 |
| Gad1-F-200140 | 4058 | GadMARCM-F000656_seg001 | Gad1-Gal4 | F | 73297.61 | 0.46 |
| Cha-F-100299 | 4130 | ChaMARCM-F001385_seg001 | Cha-Gal4 | F | 2216.68 | 0.35 |
| Cha-F-300264 | 5690 | ChaMARCM-F000948_seg001 | Cha-Gal4 | F | 62938.84 | 0.46 |
| Cha-F-700129 | 7698 | ChaMARCM-F000516_seg001 | Cha-Gal4 | F | 119961.90 | 0.36 |
| Trh-F-700030 | 10451 | TPHMARCM-810F_seg1 | Trh-Gal4 | F | 202191.96 | 0.58 |
| VGlut-F-500468 | 12252 | DvGlutMARCM-F002714_seg001 | VGlut-Gal4 | F | 99744.44 | 0.49 |
| Trh-F-000051 | 12593 | TPHMARCM-F001389_seg001 | Trh-Gal4 | F | 236762.95 | 0.67 |
| Trh-F-400086 | 12723 | TPHMARCM-884F_seg1 | Trh-Gal4 | F | 233707.32 | 0.66 |
| VGlut-F-000432 | 12741 | DVGlutMARCM-F2001_seg1 | VGlut-Gal4 | F | 240943.85 | 0.66 |
| Trh-F-800001 | 13252 | TPHMARCM-739F_seg1 | Trh-Gal4 | F | 233741.30 | 0.66 |
| Trh-F-700024 | 13264 | TPHMARCM-752F_seg1 | Trh-Gal4 | F | 235925.90 | 0.67 |
| Trh-F-100000 | 13783 | TPHMARCM-001F_seg1 | Trh-Gal4 | F | 253229.46 | 0.69 |
| Trh-F-100012 | 13802 | TPHMARCM-020F_seg1 | Trh-Gal4 | F | 232362.60 | 0.65 |
| Trh-F-300000 | 13821 | TPHMARCM-041F_seg1 | Trh-Gal4 | F | 237366.90 | 0.66 |
| Trh-F-300009 | 13888 | TPHMARCM-153F_seg1 | Trh-Gal4 | F | 243329.93 | 0.67 |
| Trh-F-200014 | 13933 | TPHMARCM-222F_seg1 | Trh-Gal4 | F | 226565.08 | 0.66 |
| Trh-F-700008 | 14115 | TPHMARCM-475F_seg1 | Trh-Gal4 | F | 230182.29 | 0.65 |
| Trh-F-600029 | 14189 | TPHMARCM-631F_seg1 | Trh-Gal4 | F | 224022.89 | 0.66 |
| Trh-F-700016 | 14228 | TPHMARCM-680F_seg1 | Trh-Gal4 | F | 228501.77 | 0.67 |
| VGlut-F-100061 | 14501 | DvGlutMARCM-F774_seg1 | VGlut-Gal4 | F | 221540.39 | 0.66 |
| VGlut-F-300228 | 14909 | DvGlutMARCM-F1252_seg1 | VGlut-Gal4 | F | 8819.55 | 0.36 |
| VGlut-F-200249 | 14917 | DvGlutMARCM-F1259_seg1 | VGlut-Gal4 | F | 221501.07 | 0.62 |