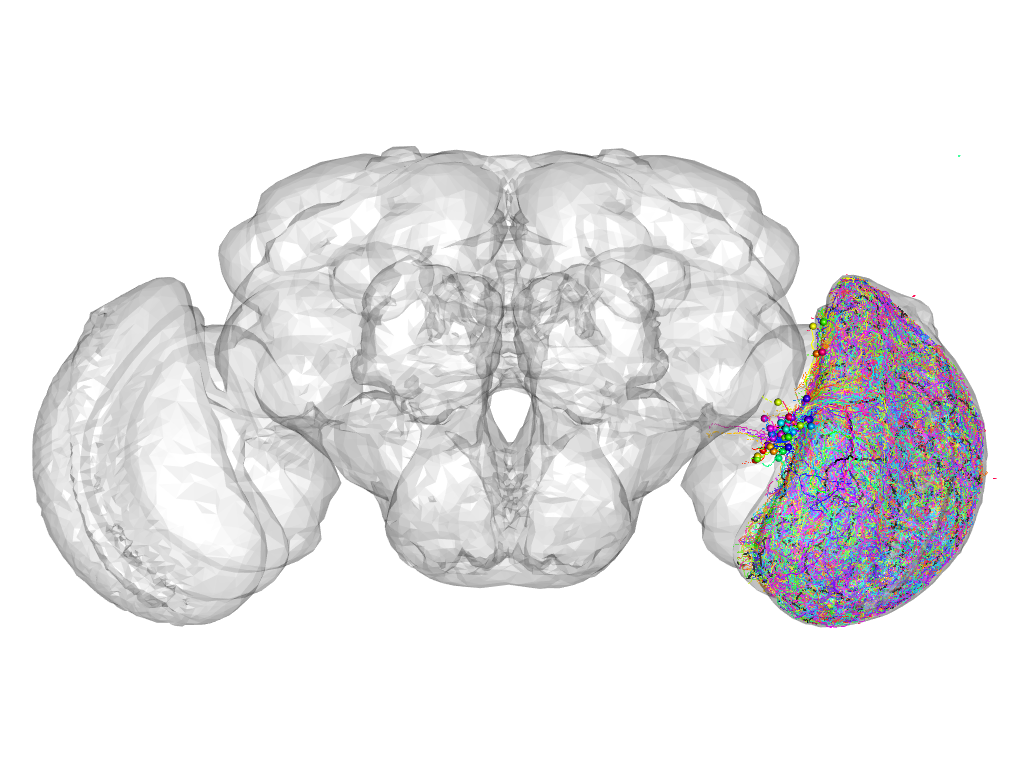
Cluster 209
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-600023), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 175 | Trh-M-700055 | III | 0.49 |
| 468 | VGlut-F-600670 | III | 0.40 |
| 759 | VGlut-F-600257 | III | 0.24 |
| 661 | Tdc2-F-000027 | VII | 0.21 |
| 660 | Tdc2-F-200049 | I | 0.18 |
Cluster composition
This cluster has 42 neurons. The exemplar of this cluster (Trh-M-600023) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-600023 | 3620 | TPHMARCM-583M_seg1 | Trh-Gal4 | M | 120691.67 | 1.00 |
| Trh-M-000141 | 1733 | TPHMARCM-M001810_seg001 | Trh-Gal4 | M | 59164.54 | 0.57 |
| Trh-M-000143 | 1735 | TPHMARCM-M001812_seg001 | Trh-Gal4 | M | 68153.39 | 0.60 |
| Trh-M-000146 | 1738 | TPHMARCM-M001815_seg001 | Trh-Gal4 | M | 68230.52 | 0.61 |
| Trh-M-000019 | 1977 | TPHMARCM-0353M_seg001 | Trh-Gal4 | M | 73762.13 | 0.63 |
| Trh-M-000067 | 2214 | TPHMARCM-M001705_seg001 | Trh-Gal4 | M | 65452.49 | 0.60 |
| Trh-M-000081 | 2251 | TPHMARCM-M001738_seg001 | Trh-Gal4 | M | 65622.09 | 0.58 |
| Trh-M-000114 | 2300 | TPHMARCM-M001782_seg001 | Trh-Gal4 | M | 72408.74 | 0.64 |
| Trh-M-100072 | 2621 | TPHMARCM-M001355_seg001 | Trh-Gal4 | M | 72405.39 | 0.62 |
| Trh-M-000035 | 2673 | TPHMARCM-M001439_seg001 | Trh-Gal4 | M | 81650.90 | 0.67 |
| Trh-M-000040 | 2685 | TPHMARCM-M001452_seg001 | Trh-Gal4 | M | 65389.93 | 0.59 |
| Trh-M-800020 | 2750 | TPHMARCM-M001516_seg001 | Trh-Gal4 | M | 63930.87 | 0.60 |
| Trh-M-300100 | 2855 | TPHMARCM-1083M_seg1 | Trh-Gal4 | M | 62725.00 | 0.57 |
| Trh-M-600064 | 2888 | TPHMARCM-1128M_seg3 | Trh-Gal4 | M | 51805.98 | 0.53 |
| Trh-M-900019 | 2956 | TPHMARCM-897M_seg1 | Trh-Gal4 | M | 63645.46 | 0.59 |
| Trh-M-600047 | 3006 | TPHMARCM-951M_seg1 | Trh-Gal4 | M | 70257.98 | 0.63 |
| Trh-M-700019 | 3017 | TPHMARCM-969M_seg1 | Trh-Gal4 | M | 74324.97 | 0.65 |
| Trh-M-100009 | 3391 | TPHMARCM-073M_seg1 | Trh-Gal4 | M | 64212.36 | 0.59 |
| Trh-M-100018 | 3407 | TPHMARCM-092M_seg1 | Trh-Gal4 | M | 83562.30 | 0.69 |
| Trh-M-400000 | 3430 | TPHMARCM-173M_seg1 | Trh-Gal4 | M | 71650.13 | 0.62 |
| Trh-M-900003 | 3453 | TPHMARCM-210M_seg1 | Trh-Gal4 | M | 64949.48 | 0.61 |
| Trh-M-000013 | 3476 | TPHMARCM-347M_seg1 | Trh-Gal4 | M | 65098.48 | 0.58 |
| Trh-M-100039 | 3551 | TPHMARCM-482M_seg1 | Trh-Gal4 | M | 82026.61 | 0.68 |
| Trh-M-500024 | 3562 | TPHMARCM-494M_seg1 | Trh-Gal4 | M | 69327.99 | 0.61 |
| Trh-M-100048 | 3577 | TPHMARCM-507M_seg1 | Trh-Gal4 | M | 74267.36 | 0.64 |
| Trh-M-500029 | 3584 | TPHMARCM-527M_seg1 | Trh-Gal4 | M | 79526.63 | 0.66 |
| Trh-M-500033 | 3616 | TPHMARCM-574M_seg1 | Trh-Gal4 | M | 56414.39 | 0.55 |
| Trh-M-800002 | 3631 | TPHMARCM-604M_seg1 | Trh-Gal4 | M | 75887.17 | 0.66 |
| Trh-M-600027 | 3637 | TPHMARCM-627M_seg1 | Trh-Gal4 | M | 81208.24 | 0.68 |
| VGlut-F-300541 | 7992 | DvGlutMARCM-F003814_seg001 | VGlut-Gal4 | F | 30192.92 | 0.46 |
| Trh-F-000065 | 10045 | TPHMARCM-F001827_seg001 | Trh-Gal4 | F | 58670.14 | 0.56 |
| Trh-F-000092 | 10070 | TPHMARCM-F001851_seg001 | Trh-Gal4 | F | 66408.26 | 0.59 |
| Trh-F-000056 | 10456 | TPHMARCM-F001401_seg001 | Trh-Gal4 | F | 80569.25 | 0.66 |
| Trh-F-000047 | 12569 | TPHMARCM-F001362_seg001 | Trh-Gal4 | F | 81348.62 | 0.67 |
| Trh-F-100002 | 13785 | TPHMARCM-003F_seg1 | Trh-Gal4 | F | 81905.28 | 0.67 |
| Trh-F-100009 | 13792 | TPHMARCM-010F_seg1 | Trh-Gal4 | F | 83197.78 | 0.68 |
| Trh-F-000019 | 13917 | TPHMARCM-200F_seg1 | Trh-Gal4 | F | 74545.53 | 0.65 |
| Trh-F-200015 | 13934 | TPHMARCM-223F_seg1 | Trh-Gal4 | F | 79613.55 | 0.67 |
| Trh-F-200035 | 13966 | TPHMARCM-256F_seg1 | Trh-Gal4 | F | 66059.84 | 0.61 |
| Trh-F-700009 | 14116 | TPHMARCM-476F_seg1 | Trh-Gal4 | F | 70735.81 | 0.63 |
| VGlut-F-900012 | 14892 | DvGlutMARCM-F1236_seg2 | VGlut-Gal4 | F | 80445.86 | 0.65 |
| VGlut-F-000064 | 15990 | DvGlutMARCM-F379_seg1 | VGlut-Gal4 | F | -5916.90 | 0.33 |