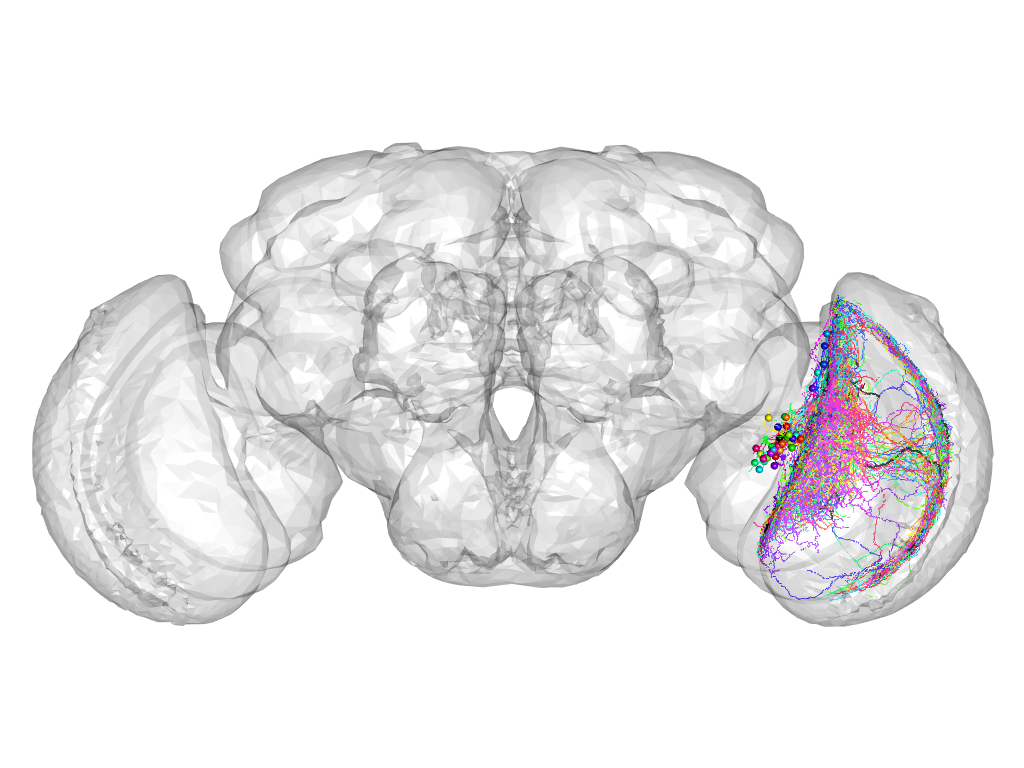
Cluster 1040
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-200092), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 661 | Tdc2-F-000027 | VII | 0.74 |
| 1020 | VGlut-F-100029 | III | 0.72 |
| 173 | Trh-M-000051 | III | 0.71 |
| 16 | fru-M-300276 | III | 0.67 |
| 952 | VGlut-F-600052 | III | 0.63 |
| 1006 | VGlut-F-500043 | III | 0.62 |
| 759 | VGlut-F-600257 | III | 0.60 |
| 874 | Trh-F-100011 | III | 0.59 |
| 76 | fru-M-300240 | III | 0.54 |
| 919 | VGlut-F-000205 | III | 0.53 |
| 468 | VGlut-F-600670 | III | 0.52 |
Cluster composition
This cluster has 38 neurons. The exemplar of this cluster (VGlut-F-200092) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-200092 | 16108 | DvGlutMARCM-F491_seg1 | VGlut-Gal4 | F | 18234.16 | 1.00 |
| fru-M-000194 | 303 | FruMARCM-M002530_seg001 | fru-Gal4 | M | 12674.34 | 0.70 |
| fru-M-700137 | 916 | FruMARCM-M002177_seg001 | fru-Gal4 | M | 11447.32 | 0.64 |
| fru-F-800064 | 3765 | FruMARCM-F002359_seg001 | fru-Gal4 | F | 12142.43 | 0.68 |
| fru-F-200166 | 3786 | FruMARCM-F002460_seg001 | fru-Gal4 | F | 11580.00 | 0.64 |
| fru-F-200108 | 4555 | FruMARCM-F001665_seg001 | fru-Gal4 | F | 11738.71 | 0.66 |
| VGlut-F-500865 | 5795 | DvGlutMARCM-F004494_seg001 | VGlut-Gal4 | F | 11604.99 | 0.67 |
| VGlut-F-600779 | 6423 | DvGlutMARCM-F004216_seg001 | VGlut-Gal4 | F | 10799.11 | 0.62 |
| VGlut-F-400884 | 6457 | DvGlutMARCM-F004242_seg001 | VGlut-Gal4 | F | 12104.94 | 0.70 |
| VGlut-F-700273 | 8509 | DvGlutMARCM-F003092_seg001 | VGlut-Gal4 | F | 12132.64 | 0.67 |
| VGlut-F-600524 | 8553 | DvGlutMARCM-F003122_seg001 | VGlut-Gal4 | F | 11857.90 | 0.66 |
| VGlut-F-700308 | 8581 | DvGlutMARCM-F003144_seg001 | VGlut-Gal4 | F | 11092.16 | 0.64 |
| VGlut-F-200510 | 8835 | DvGlutMARCM-F003633_seg001 | VGlut-Gal4 | F | 12516.88 | 0.68 |
| VGlut-F-700364 | 8919 | DvGlutMARCM-F003496_seg001 | VGlut-Gal4 | F | 11067.70 | 0.65 |
| VGlut-F-500666 | 9084 | DvGlutMARCM-F003399_seg001 | VGlut-Gal4 | F | 12558.06 | 0.69 |
| VGlut-F-000494 | 9226 | DvGlutMARCM-F003290_seg001 | VGlut-Gal4 | F | 10470.39 | 0.60 |
| VGlut-F-200458 | 9321 | DvGlutMARCM-F003159_seg001 | VGlut-Gal4 | F | 10391.55 | 0.60 |
| VGlut-F-400681 | 9416 | DvGlutMARCM-F003232_seg001 | VGlut-Gal4 | F | 11588.18 | 0.65 |
| VGlut-F-300212 | 10224 | DvGlutMARCM-F1122_seg1 | VGlut-Gal4 | F | 8028.80 | 0.50 |
| VGlut-F-200435 | 11025 | DvGlutMARCM-F002907_seg001 | VGlut-Gal4 | F | 12186.20 | 0.66 |
| VGlut-F-300428 | 11047 | DvGlutMARCM-F002927_seg001 | VGlut-Gal4 | F | 11518.32 | 0.64 |
| VGlut-F-800114 | 11149 | DvGlutMARCM-F003003_seg001 | VGlut-Gal4 | F | 12160.25 | 0.67 |
| VGlut-F-400647 | 11673 | DvGlutMARCM-F002249_seg001 | VGlut-Gal4 | F | 11942.63 | 0.66 |
| VGlut-F-400650 | 11676 | DvGlutMARCM-F002252_seg001 | VGlut-Gal4 | F | 13235.14 | 0.70 |
| VGlut-F-300402 | 11725 | DvGlutMARCM-F002298_seg001 | VGlut-Gal4 | F | 11875.68 | 0.64 |
| VGlut-F-000459 | 11846 | DvGlutMARCM-F002399_seg001 | VGlut-Gal4 | F | 11517.56 | 0.66 |
| VGlut-F-000460 | 11906 | DvGlutMARCM-F002442_seg001 | VGlut-Gal4 | F | 8817.93 | 0.51 |
| VGlut-F-300375 | 12963 | DvGlutMARCM-F1994_seg1 | VGlut-Gal4 | F | 11804.68 | 0.65 |
| VGlut-F-600035 | 14490 | DvGlutMARCM-F763_seg1 | VGlut-Gal4 | F | 11804.43 | 0.64 |
| VGlut-F-100066 | 14621 | DvGlutMARCM-F879_seg1 | VGlut-Gal4 | F | 9577.29 | 0.48 |
| VGlut-F-200238 | 14849 | DvGlutMARCM-F1196_seg1 | VGlut-Gal4 | F | 9366.71 | 0.57 |
| VGlut-F-500151 | 14948 | DvGlutMARCM-F1286_seg1 | VGlut-Gal4 | F | 12335.95 | 0.67 |
| VGlut-F-300264 | 15143 | DvGlutMARCM-F1476_seg1 | VGlut-Gal4 | F | 11398.28 | 0.60 |
| VGlut-F-100048 | 15446 | DvGlutMARCM-F604_seg1 | VGlut-Gal4 | F | 12655.31 | 0.69 |
| VGlut-F-200043 | 15661 | DvGlutMARCM-F074_seg1 | VGlut-Gal4 | F | 11450.82 | 0.62 |
| VGlut-F-900001 | 15793 | DvGlutMARCM-F206_seg1 | VGlut-Gal4 | F | 12688.03 | 0.67 |
| VGlut-F-500051 | 15869 | DvGlutMARCM-F272_seg1 | VGlut-Gal4 | F | 11943.95 | 0.62 |
| VGlut-F-300082 | 16127 | DvGlutMARCM-F510_seg1 | VGlut-Gal4 | F | 12663.50 | 0.66 |