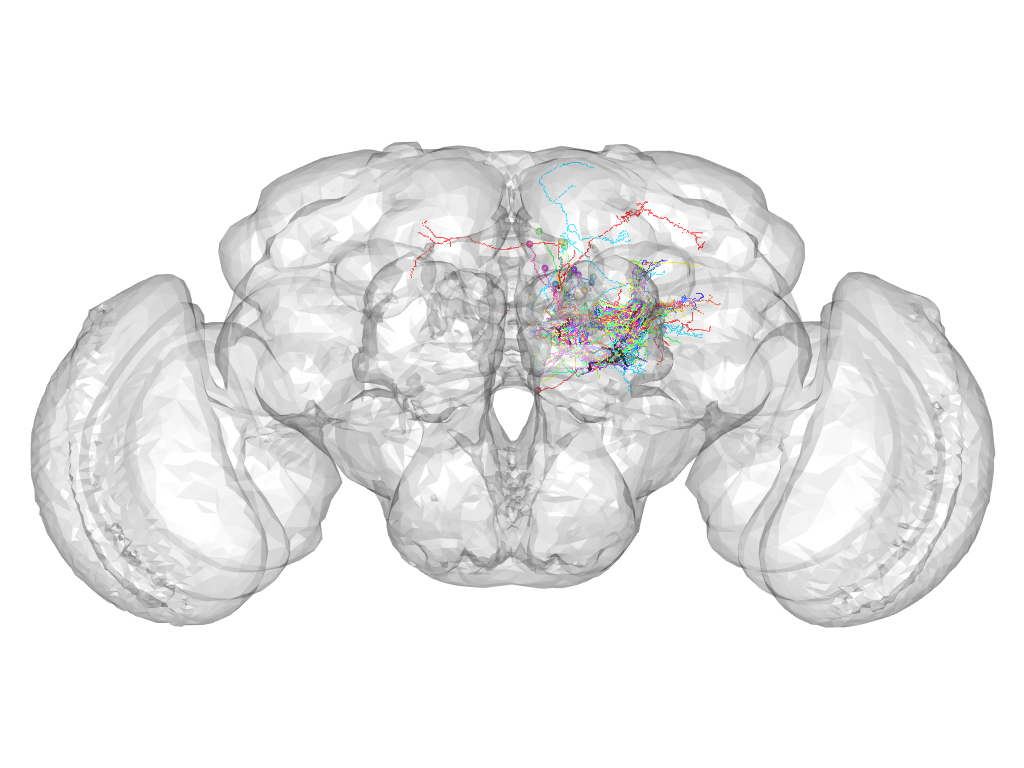
Cluster 447
Part of supercluster VI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-400843), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 666 | TH-F-200011 | IX | 0.75 |
| 872 | TH-F-100101 | IX | 0.73 |
| 915 | VGlut-F-400204 | VI | 0.70 |
| 707 | VGlut-F-200430 | VII | 0.70 |
| 425 | VGlut-F-500805 | VII | 0.68 |
| 683 | VGlut-F-000282 | VI | 0.67 |
| 380 | VGlut-F-200603 | VI | 0.65 |
| 179 | Trh-M-100061 | VI | 0.65 |
| 873 | TH-F-000089 | IX | 0.62 |
| 406 | fru-F-000053 | VII | 0.62 |
| 353 | Cha-F-200264 | VI | 0.62 |
| 964 | VGlut-F-200325 | XIV | 0.61 |
| 904 | VGlut-F-100092 | XVII | 0.61 |
| 1033 | VGlut-F-000081 | XVIII | 0.57 |
| 956 | VGlut-F-300289 | VI | 0.53 |
| 629 | fru-F-100015 | VII | 0.51 |
| 1045 | VGlut-F-300086 | VI | 0.51 |
| 826 | VGlut-F-500316 | VI | 0.51 |
| 686 | VGlut-F-900065 | VII | 0.50 |
Cluster composition
This cluster has 20 neurons. The exemplar of this cluster (VGlut-F-400843) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-400843 | 6864 | DvGlutMARCM-F004031_seg001 | VGlut-Gal4 | F | 11047.55 | 1.00 |
| fru-M-200069 | 1577 | FruMARCM-M000646_seg003 | fru-Gal4 | M | 6023.72 | 0.29 |
| VGlut-F-700118 | 6060 | DvGlutMARCM-F002772_seg003 | VGlut-Gal4 | F | 7558.03 | 0.66 |
| VGlut-F-700525 | 6450 | DvGlutMARCM-F004235_seg001 | VGlut-Gal4 | F | 7446.84 | 0.71 |
| VGlut-F-500798 | 6520 | DvGlutMARCM-F004293_seg001 | VGlut-Gal4 | F | 7789.33 | 0.61 |
| VGlut-F-500807 | 6547 | DvGlutMARCM-F004310_seg001 | VGlut-Gal4 | F | 7385.57 | 0.62 |
| VGlut-F-300643 | 6620 | DvGlutMARCM-F004373_seg001 | VGlut-Gal4 | F | 7090.40 | 0.69 |
| VGlut-F-700489 | 7019 | DvGlutMARCM-F004156_seg002 | VGlut-Gal4 | F | 7842.14 | 0.68 |
| VGlut-F-700490 | 7020 | DvGlutMARCM-F004156_seg003 | VGlut-Gal4 | F | 7857.35 | 0.67 |
| VGlut-F-700512 | 7054 | DvGlutMARCM-F004182_seg002 | VGlut-Gal4 | F | 7551.46 | 0.68 |
| VGlut-F-700291 | 8528 | DvGlutMARCM-F003106_seg001 | VGlut-Gal4 | F | 7293.93 | 0.63 |
| Gad1-F-700034 | 9675 | GadMARCM-F000526_seg001 | Gad1-Gal4 | F | 6499.07 | 0.35 |
| VGlut-F-600404 | 10781 | DvGlutMARCM-F002730_seg002 | VGlut-Gal4 | F | 8064.72 | 0.73 |
| VGlut-F-500480 | 11005 | DvGlutMARCM-F002891_seg001 | VGlut-Gal4 | F | 7627.45 | 0.63 |
| VGlut-F-600500 | 11651 | DvGlutMARCM-F002225_seg001 | VGlut-Gal4 | F | 7320.08 | 0.64 |
| VGlut-F-600221 | 11821 | DvGlutMARCM-F002385_seg001 | VGlut-Gal4 | F | 7575.12 | 0.71 |
| VGlut-F-400424 | 12752 | DvGlutMARCM-F1811_seg1 | VGlut-Gal4 | F | 8199.31 | 0.70 |
| VGlut-F-400299 | 14360 | DvGlutMARCM-F1041_seg2 | VGlut-Gal4 | F | 6313.34 | 0.60 |
| VGlut-F-600029 | 14481 | DvGlutMARCM-F759-x3_seg2 | VGlut-Gal4 | F | 7232.89 | 0.61 |
| VGlut-F-400158 | 15575 | DvGlutMARCM-F703_seg1 | VGlut-Gal4 | F | 7345.13 | 0.65 |