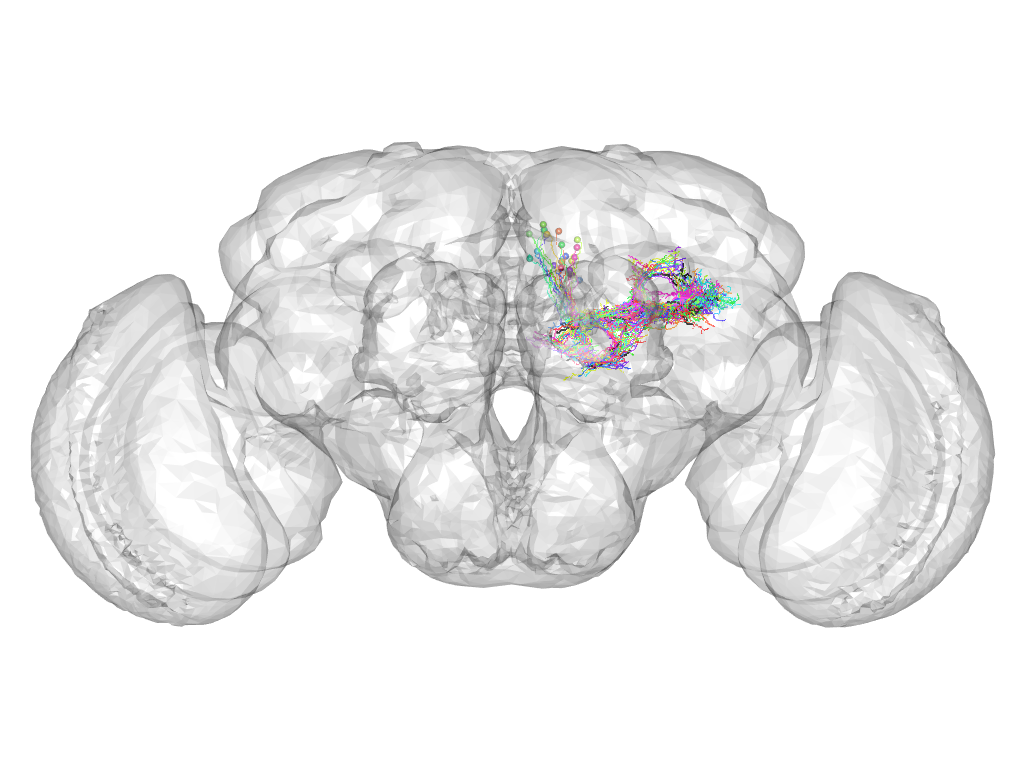
Cluster 915
Part of supercluster VI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-400204), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 666 | TH-F-200011 | IX | 0.77 |
| 872 | TH-F-100101 | IX | 0.70 |
| 904 | VGlut-F-100092 | XVII | 0.56 |
| 873 | TH-F-000089 | IX | 0.53 |
| 1033 | VGlut-F-000081 | XVIII | 0.51 |
| 1014 | VGlut-F-100022 | XVII | 0.50 |
Cluster composition
This cluster has 23 neurons. The exemplar of this cluster (VGlut-F-400204) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-400204 | 14484 | DvGlutMARCM-F760-x2_seg1 | VGlut-Gal4 | F | 8633.04 | 1.00 |
| VGlut-F-400881 | 6425 | DvGlutMARCM-F004218_seg001 | VGlut-Gal4 | F | 5641.53 | 0.67 |
| VGlut-F-800277 | 6544 | DvGlutMARCM-F004308_seg002 | VGlut-Gal4 | F | 5981.50 | 0.68 |
| VGlut-F-500811 | 6561 | DvGlutMARCM-F004323_seg001 | VGlut-Gal4 | F | 5390.83 | 0.63 |
| VGlut-F-400836 | 6857 | DvGlutMARCM-F004029_seg004 | VGlut-Gal4 | F | 5182.59 | 0.63 |
| VGlut-F-800254 | 6954 | DvGlutMARCM-F004107_seg002 | VGlut-Gal4 | F | 5255.54 | 0.62 |
| VGlut-F-800120 | 10794 | DvGlutMARCM-F003028_seg001 | VGlut-Gal4 | F | 5751.25 | 0.65 |
| VGlut-F-600434 | 10926 | DvGlutMARCM-F002822_seg002 | VGlut-Gal4 | F | 5535.67 | 0.64 |
| VGlut-F-600249 | 11853 | DvGlutMARCM-F002403_seg003 | VGlut-Gal4 | F | 5658.31 | 0.65 |
| VGlut-F-800065 | 12116 | DvGlutMARCM-F002613_seg001 | VGlut-Gal4 | F | 5641.56 | 0.66 |
| VGlut-F-600346 | 12175 | DvGlutMARCM-F002655_seg002 | VGlut-Gal4 | F | 5627.48 | 0.65 |
| VGlut-F-400425 | 12753 | DvGlutMARCM-F1812_seg1 | VGlut-Gal4 | F | 5520.47 | 0.63 |
| VGlut-F-500307 | 12888 | DvGlutMARCM-F1930_seg1 | VGlut-Gal4 | F | 5396.37 | 0.65 |
| VGlut-F-300357 | 12895 | DvGlutMARCM-F1938_seg1 | VGlut-Gal4 | F | 5726.80 | 0.67 |
| VGlut-F-600114 | 12947 | DvGlutMARCM-F1980_seg2 | VGlut-Gal4 | F | 5765.27 | 0.69 |
| VGlut-F-600120 | 13048 | DvGlutMARCM-F2071_seg1 | VGlut-Gal4 | F | 5301.52 | 0.65 |
| VGlut-F-600133 | 13126 | DvGlutMARCM-F2152_seg1 | VGlut-Gal4 | F | 5260.85 | 0.62 |
| VGlut-F-400285 | 14331 | DvGlutMARCM-F1013_seg2 | VGlut-Gal4 | F | 5857.27 | 0.69 |
| VGlut-F-400381 | 15094 | DvGlutMARCM-F1430_seg1 | VGlut-Gal4 | F | 5918.75 | 0.69 |
| VGlut-F-200127 | 15471 | DvGlutMARCM-F629-X3_seg3 | VGlut-Gal4 | F | 5411.34 | 0.64 |
| VGlut-F-500004 | 15600 | DvGlutMARCM-F016-2singles_seg1 | VGlut-Gal4 | F | 5358.35 | 0.58 |
| VGlut-F-500050 | 15863 | DvGlutMARCM-F266_seg1 | VGlut-Gal4 | F | 5464.41 | 0.65 |
| VGlut-F-500099 | 16087 | DvGlutMARCM-F470_seg2 | VGlut-Gal4 | F | 5856.67 | 0.68 |