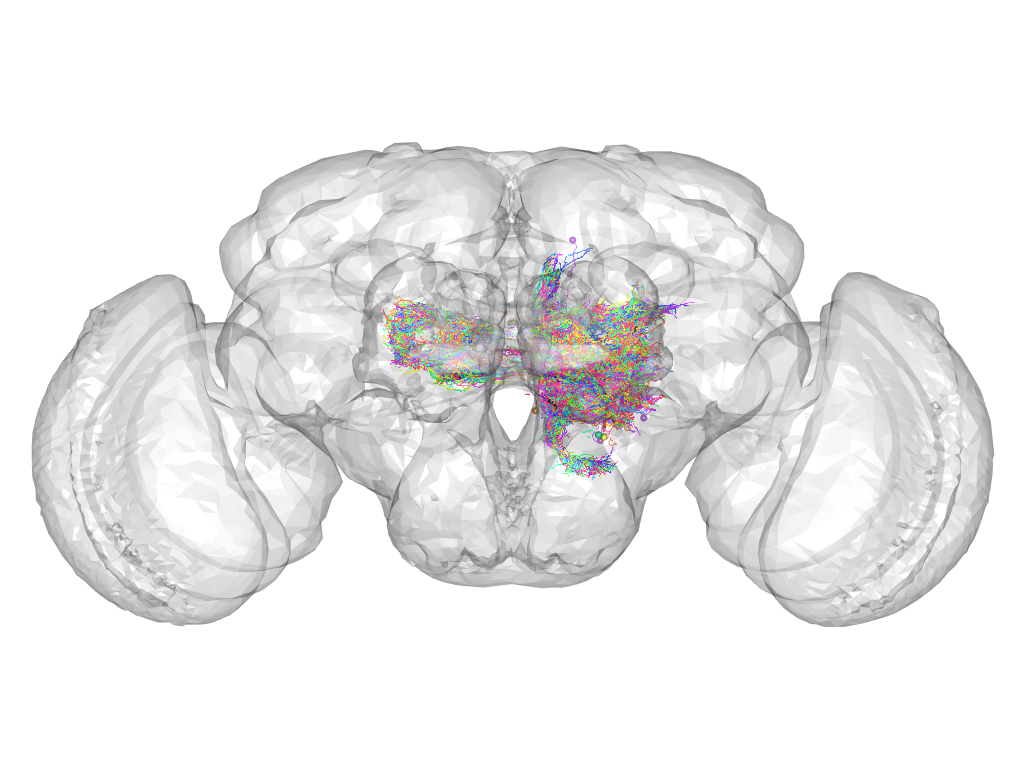
Cluster 707
Part of supercluster VII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-200430), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 425 | VGlut-F-500805 | VII | 0.77 |
| 964 | VGlut-F-200325 | XIV | 0.54 |
| 858 | TH-F-300066 | VII | 0.40 |
| 34 | fru-M-000211 | VII | 0.40 |
| 442 | VGlut-F-400823 | VII | 0.40 |
Cluster composition
This cluster has 132 neurons. The exemplar of this cluster (VGlut-F-200430) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-200430 | 10989 | DvGlutMARCM-F002878_seg001 | VGlut-Gal4 | F | 44862.18 | 1.00 |
| fru-M-200335 | 11 | FruMARCM-M002590_seg002 | fru-Gal4 | M | 32337.23 | 0.74 |
| fru-M-300249 | 22 | FruMARCM-M002216_seg001 | fru-Gal4 | M | 34498.66 | 0.77 |
| fru-M-700148 | 77 | FruMARCM-M002276_seg001 | fru-Gal4 | M | 33123.49 | 0.74 |
| fru-M-800094 | 84 | FruMARCM-M002283_seg001 | fru-Gal4 | M | 33982.40 | 0.75 |
| fru-M-900057 | 111 | FruMARCM-M002309_seg001 | fru-Gal4 | M | 33468.84 | 0.76 |
| fru-M-300280 | 215 | FruMARCM-M002431_seg001 | fru-Gal4 | M | 33669.96 | 0.75 |
| fru-M-200305 | 298 | FruMARCM-M002525_seg002 | fru-Gal4 | M | 33388.36 | 0.74 |
| fru-M-400228 | 397 | FruMARCM-M002629_seg001 | fru-Gal4 | M | 22711.70 | 0.59 |
| fru-M-400177 | 417 | FruMARCM-M001935_seg001 | fru-Gal4 | M | 21835.78 | 0.57 |
| fru-M-000095 | 564 | FruMARCM-M001739_seg001 | fru-Gal4 | M | 33165.78 | 0.75 |
| fru-M-200222 | 642 | FruMARCM-M001850_seg001 | fru-Gal4 | M | 34036.91 | 0.76 |
| fru-M-500196 | 742 | FruMARCM-M001957_seg001 | fru-Gal4 | M | 33408.16 | 0.76 |
| fru-M-100192 | 860 | FruMARCM-M002117_seg001 | fru-Gal4 | M | 33840.58 | 0.76 |
| fru-M-500219 | 870 | FruMARCM-M002124_seg001 | fru-Gal4 | M | 34795.37 | 0.76 |
| fru-M-100204 | 915 | FruMARCM-M002176_seg001 | fru-Gal4 | M | 34695.55 | 0.77 |
| fru-M-500104 | 987 | FruMARCM-M001169_seg001 | fru-Gal4 | M | 34256.03 | 0.76 |
| fru-M-700081 | 1217 | FruMARCM-M001402_seg001 | fru-Gal4 | M | 33264.99 | 0.74 |
| fru-M-100067 | 1649 | FruMARCM-M000789_seg001 | fru-Gal4 | M | 33920.79 | 0.75 |
| fru-M-200060 | 1935 | FruMARCM-M000533_seg001 | fru-Gal4 | M | 32468.77 | 0.74 |
| fru-M-500033 | 1956 | FruMARCM-M000599_seg001 | fru-Gal4 | M | 34133.53 | 0.76 |
| Trh-M-200103 | 2203 | TPHMARCM-M001692_seg002 | Trh-Gal4 | M | 21509.70 | 0.60 |
| Trh-M-300041 | 3540 | TPHMARCM-445M_seg2 | Trh-Gal4 | M | 23897.05 | 0.62 |
| fru-F-200164 | 3777 | FruMARCM-F002405_seg001 | fru-Gal4 | F | 34764.83 | 0.76 |
| Gad1-F-900039 | 3964 | GadMARCM-F000588_seg001 | Gad1-Gal4 | F | 29574.26 | 0.70 |
| fru-F-000082 | 4553 | FruMARCM-F001661_seg001 | fru-Gal4 | F | 33413.27 | 0.74 |
| VGlut-F-400894 | 5695 | DvGlutMARCM-F004444_seg001 | VGlut-Gal4 | F | 33690.48 | 0.75 |
| VGlut-F-600792 | 5720 | DvGlutMARCM-F004427_seg001 | VGlut-Gal4 | F | 34641.12 | 0.77 |
| VGlut-F-400893 | 5736 | DvGlutMARCM-F004442_seg001 | VGlut-Gal4 | F | 33579.30 | 0.76 |
| VGlut-F-200595 | 5740 | DvGlutMARCM-F004447_seg001 | VGlut-Gal4 | F | 33941.90 | 0.77 |
| VGlut-F-400897 | 5747 | DvGlutMARCM-F004452_seg001 | VGlut-Gal4 | F | 33876.09 | 0.75 |
| VGlut-F-900135 | 5801 | DvGlutMARCM-F004499_seg001 | VGlut-Gal4 | F | 33817.28 | 0.76 |
| VGlut-F-400907 | 5807 | DvGlutMARCM-F004504_seg001 | VGlut-Gal4 | F | 33426.32 | 0.74 |
| VGlut-F-900139 | 5890 | DvGlutMARCM-F004569_seg001 | VGlut-Gal4 | F | 34143.02 | 0.76 |
| VGlut-F-600136 | 5979 | DvGlutMARCM-F2158_seg1 | VGlut-Gal4 | F | 33458.82 | 0.76 |
| VGlut-F-900043 | 6047 | DvGlutMARCM-F002754_seg002 | VGlut-Gal4 | F | 33777.13 | 0.74 |
| VGlut-F-700524 | 6449 | DvGlutMARCM-F004234_seg001 | VGlut-Gal4 | F | 33642.55 | 0.76 |
| VGlut-F-300621 | 6484 | DvGlutMARCM-F004262_seg001 | VGlut-Gal4 | F | 33965.60 | 0.75 |
| VGlut-F-800264 | 6509 | DvGlutMARCM-F004287_seg001 | VGlut-Gal4 | F | 33830.66 | 0.75 |
| VGlut-F-300644 | 6636 | DvGlutMARCM-F004384_seg001 | VGlut-Gal4 | F | 34107.62 | 0.77 |
| VGlut-F-000613 | 6678 | DvGlutMARCM-F003900_seg001 | VGlut-Gal4 | F | 33886.08 | 0.76 |
| VGlut-F-300569 | 6793 | DvGlutMARCM-F003986_seg001 | VGlut-Gal4 | F | 33216.25 | 0.75 |
| VGlut-F-000623 | 6890 | DvGlutMARCM-F004046_seg001 | VGlut-Gal4 | F | 33691.18 | 0.75 |
| VGlut-F-200551 | 6927 | DvGlutMARCM-F004082_seg001 | VGlut-Gal4 | F | 34066.01 | 0.76 |
| VGlut-F-100360 | 6951 | DvGlutMARCM-F004105_seg001 | VGlut-Gal4 | F | 33854.84 | 0.75 |
| VGlut-F-100362 | 6974 | DvGlutMARCM-F004120_seg001 | VGlut-Gal4 | F | 34572.66 | 0.76 |
| VGlut-F-000641 | 6986 | DvGlutMARCM-F004132_seg001 | VGlut-Gal4 | F | 34773.24 | 0.77 |
| VGlut-F-000642 | 6987 | DvGlutMARCM-F004133_seg001 | VGlut-Gal4 | F | 35155.12 | 0.78 |
| VGlut-F-400877 | 7003 | DvGlutMARCM-F004144_seg001 | VGlut-Gal4 | F | 33916.38 | 0.75 |
| VGlut-F-600766 | 7008 | DvGlutMARCM-F004149_seg001 | VGlut-Gal4 | F | 33466.98 | 0.73 |
| VGlut-F-300595 | 7016 | DvGlutMARCM-F004154_seg002 | VGlut-Gal4 | F | 33621.93 | 0.76 |
| VGlut-F-500776 | 7068 | DvGlutMARCM-F004197_seg001 | VGlut-Gal4 | F | 33636.56 | 0.72 |
| VGlut-F-600698 | 7418 | DvGlutMARCM-F003875_seg001 | VGlut-Gal4 | F | 33235.92 | 0.75 |
| VGlut-F-000602 | 7981 | DvGlutMARCM-F003806_seg001 | VGlut-Gal4 | F | 32854.53 | 0.75 |
| VGlut-F-200539 | 8000 | DvGlutMARCM-F003820_seg001 | VGlut-Gal4 | F | 32528.14 | 0.74 |
| VGlut-F-100343 | 8021 | DvGlutMARCM-F003836_seg001 | VGlut-Gal4 | F | 34927.70 | 0.77 |
| VGlut-F-100344 | 8022 | DvGlutMARCM-F003837_seg001 | VGlut-Gal4 | F | 34114.26 | 0.76 |
| VGlut-F-300447 | 8463 | DvGlutMARCM-F003061_seg001 | VGlut-Gal4 | F | 33516.90 | 0.76 |
| VGlut-F-700271 | 8507 | DvGlutMARCM-F003090_seg001 | VGlut-Gal4 | F | 32578.81 | 0.75 |
| VGlut-F-000574 | 8668 | DvGlutMARCM-F003715_seg001 | VGlut-Gal4 | F | 34830.66 | 0.76 |
| VGlut-F-400760 | 8670 | DvGlutMARCM-F003717_seg001 | VGlut-Gal4 | F | 33294.73 | 0.74 |
| VGlut-F-300527 | 8701 | DvGlutMARCM-F003734_seg001 | VGlut-Gal4 | F | 34417.48 | 0.77 |
| VGlut-F-000542 | 8766 | DvGlutMARCM-F003587_seg001 | VGlut-Gal4 | F | 32287.44 | 0.74 |
| VGlut-F-200511 | 8854 | DvGlutMARCM-F003644_seg001 | VGlut-Gal4 | F | 33379.73 | 0.76 |
| VGlut-F-300511 | 8861 | DvGlutMARCM-F003650_seg001 | VGlut-Gal4 | F | 32660.77 | 0.75 |
| VGlut-F-800206 | 8909 | DvGlutMARCM-F003483_seg001 | VGlut-Gal4 | F | 33398.09 | 0.72 |
| VGlut-F-400725 | 9006 | DvGlutMARCM-F003550_seg001 | VGlut-Gal4 | F | 33901.72 | 0.76 |
| VGlut-F-000503 | 9025 | DvGlutMARCM-F003349_seg001 | VGlut-Gal4 | F | 33474.18 | 0.76 |
| VGlut-F-000510 | 9051 | DvGlutMARCM-F003373_seg001 | VGlut-Gal4 | F | 32723.81 | 0.75 |
| VGlut-F-000512 | 9085 | DvGlutMARCM-F003400_seg001 | VGlut-Gal4 | F | 33288.87 | 0.74 |
| VGlut-F-600610 | 9138 | DvGlutMARCM-F003430_seg001 | VGlut-Gal4 | F | 33972.28 | 0.77 |
| VGlut-F-500614 | 9194 | DvGlutMARCM-F003262_seg001 | VGlut-Gal4 | F | 34052.77 | 0.76 |
| VGlut-F-800142 | 9240 | DvGlutMARCM-F003303_seg001 | VGlut-Gal4 | F | 33411.09 | 0.75 |
| VGlut-F-500634 | 9257 | DvGlutMARCM-F003316_seg001 | VGlut-Gal4 | F | 33581.75 | 0.75 |
| VGlut-F-300449 | 9303 | DvGlutMARCM-F003150_seg001 | VGlut-Gal4 | F | 33195.01 | 0.75 |
| VGlut-F-300470 | 9326 | DvGlutMARCM-F003164_seg001 | VGlut-Gal4 | F | 33949.54 | 0.75 |
| VGlut-F-500587 | 9352 | DvGlutMARCM-F003184_seg001 | VGlut-Gal4 | F | 33167.57 | 0.75 |
| VGlut-F-700146 | 10871 | DvGlutMARCM-F002783_seg001 | VGlut-Gal4 | F | 34723.79 | 0.77 |
| VGlut-F-700207 | 10942 | DvGlutMARCM-F002835_seg001 | VGlut-Gal4 | F | 33688.13 | 0.76 |
| VGlut-F-000468 | 10987 | DvGlutMARCM-F002876_seg001 | VGlut-Gal4 | F | 34408.75 | 0.76 |
| VGlut-F-200431 | 10992 | DvGlutMARCM-F002882_seg001 | VGlut-Gal4 | F | 34367.86 | 0.77 |
| VGlut-F-800092 | 11007 | DvGlutMARCM-F002892_seg002 | VGlut-Gal4 | F | 33996.43 | 0.76 |
| VGlut-F-500481 | 11009 | DvGlutMARCM-F002894_seg001 | VGlut-Gal4 | F | 29743.85 | 0.69 |
| VGlut-F-700219 | 11026 | DvGlutMARCM-F002908_seg001 | VGlut-Gal4 | F | 33551.81 | 0.75 |
| VGlut-F-200440 | 11059 | DvGlutMARCM-F002937_seg001 | VGlut-Gal4 | F | 33223.77 | 0.73 |
| VGlut-F-800103 | 11103 | DvGlutMARCM-F002972_seg002 | VGlut-Gal4 | F | 34430.02 | 0.77 |
| VGlut-F-800115 | 11150 | DvGlutMARCM-F003004_seg001 | VGlut-Gal4 | F | 34300.98 | 0.76 |
| VGlut-F-100242 | 11643 | DvGlutMARCM-F002218_seg001 | VGlut-Gal4 | F | 33312.78 | 0.75 |
| VGlut-F-400651 | 11677 | DvGlutMARCM-F002253_seg001 | VGlut-Gal4 | F | 33376.29 | 0.72 |
| VGlut-F-600200 | 11764 | DvGlutMARCM-F002335_seg001 | VGlut-Gal4 | F | 32630.78 | 0.75 |
| VGlut-F-300406 | 11880 | DvGlutMARCM-F002422_seg001 | VGlut-Gal4 | F | 33853.34 | 0.76 |
| VGlut-F-600260 | 11887 | DvGlutMARCM-F002427_seg001 | VGlut-Gal4 | F | 33317.32 | 0.75 |
| VGlut-F-300411 | 11954 | DvGlutMARCM-F002487_seg001 | VGlut-Gal4 | F | 34361.47 | 0.77 |
| VGlut-F-700070 | 12071 | DvGlutMARCM-F002581_seg001 | VGlut-Gal4 | F | 33561.69 | 0.75 |
| VGlut-F-700082 | 12087 | DvGlutMARCM-F002594_seg001 | VGlut-Gal4 | F | 33376.76 | 0.75 |
| VGlut-F-800066 | 12118 | DvGlutMARCM-F002615_seg001 | VGlut-Gal4 | F | 34135.64 | 0.77 |
| VGlut-F-600336 | 12164 | DvGlutMARCM-F002650_seg001 | VGlut-Gal4 | F | 34612.51 | 0.76 |
| VGlut-F-500449 | 12193 | DvGlutMARCM-F002667_seg001 | VGlut-Gal4 | F | 34143.27 | 0.76 |
| VGlut-F-600411 | 12279 | DvGlutMARCM-F002735_seg001 | VGlut-Gal4 | F | 34249.31 | 0.77 |
| VGlut-F-600419 | 12284 | DvGlutMARCM-F002740_seg001 | VGlut-Gal4 | F | 34598.78 | 0.74 |
| VGlut-F-700114 | 12305 | DvGlutMARCM-F002766_seg001 | VGlut-Gal4 | F | 33923.24 | 0.76 |
| VGlut-F-000481 | 12317 | DvGlutMARCM-F2209_seg1 | VGlut-Gal4 | F | 33592.57 | 0.76 |
| VGlut-F-300004 | 12363 | DvGlutMARCM-F087_seg | VGlut-Gal4 | F | 34496.02 | 0.76 |
| VGlut-F-500331 | 13037 | DvGlutMARCM-F2061_seg1 | VGlut-Gal4 | F | 32984.50 | 0.74 |
| VGlut-F-600135 | 13127 | DvGlutMARCM-F2154_seg1 | VGlut-Gal4 | F | 9482.39 | 0.44 |
| VGlut-F-300392 | 13148 | DvGlutMARCM-F2180_seg1 | VGlut-Gal4 | F | 34069.35 | 0.75 |
| Trh-F-200026 | 13954 | TPHMARCM-245F_seg1 | Trh-Gal4 | F | 21009.57 | 0.59 |
| VGlut-F-300195 | 14368 | DvGlutMARCM-F1049_seg1 | VGlut-Gal4 | F | 33509.26 | 0.75 |
| VGlut-F-300139 | 14422 | DvGlutMARCM-F716_seg1 | VGlut-Gal4 | F | 21181.33 | 0.60 |
| VGlut-F-400264 | 14634 | DvGlutMARCM-F890_seg1 | VGlut-Gal4 | F | 34774.22 | 0.76 |
| VGlut-F-000246 | 14722 | DvGlutMARCM-F982_seg1 | VGlut-Gal4 | F | 34465.19 | 0.76 |
| VGlut-F-100098 | 14746 | DvGlutMARCM-F1098_seg1 | VGlut-Gal4 | F | 34928.50 | 0.77 |
| VGlut-F-100099 | 14747 | DvGlutMARCM-F1099_seg1 | VGlut-Gal4 | F | 33333.89 | 0.75 |
| VGlut-F-000313 | 14830 | DvGlutMARCM-F1172_seg1 | VGlut-Gal4 | F | 33854.45 | 0.75 |
| VGlut-F-400346 | 14927 | DvGlutMARCM-F1267_seg1 | VGlut-Gal4 | F | 34581.68 | 0.77 |
| VGlut-F-100134 | 14951 | DvGlutMARCM-F1289_seg1 | VGlut-Gal4 | F | 34754.68 | 0.77 |
| VGlut-F-200256 | 14956 | DvGlutMARCM-F1294_seg1 | VGlut-Gal4 | F | 33802.63 | 0.76 |
| VGlut-F-400365 | 15013 | DvGlutMARCM-F1353_seg1 | VGlut-Gal4 | F | 34297.18 | 0.76 |
| VGlut-F-800028 | 15028 | DvGlutMARCM-F1367_seg1 | VGlut-Gal4 | F | 33479.47 | 0.75 |
| VGlut-F-100171 | 15332 | DvGlutMARCM-F1677_seg1 | VGlut-Gal4 | F | 33964.45 | 0.74 |
| VGlut-F-400411 | 15373 | DvGlutMARCM-F1726_seg1 | VGlut-Gal4 | F | 33832.67 | 0.75 |
| VGlut-F-100060 | 15477 | DvGlutMARCM-F634_seg1 | VGlut-Gal4 | F | 32689.19 | 0.71 |
| VGlut-F-400002 | 15619 | DvGlutMARCM-F035_seg1 | VGlut-Gal4 | F | 33192.30 | 0.74 |
| VGlut-F-200049 | 15665 | DvGlutMARCM-F080_seg1 | VGlut-Gal4 | F | 33500.84 | 0.75 |
| VGlut-F-500019 | 15731 | DvGlutMARCM-F143_seg1 | VGlut-Gal4 | F | 33933.75 | 0.75 |
| VGlut-F-700011 | 15776 | DvGlutMARCM-F182_seg1 | VGlut-Gal4 | F | 34210.17 | 0.76 |
| VGlut-F-100025 | 15892 | DvGlutMARCM-F292_seg1 | VGlut-Gal4 | F | 28913.95 | 0.68 |
| VGlut-F-300046 | 15922 | DvGlutMARCM-F317_seg1 | VGlut-Gal4 | F | 34130.44 | 0.76 |
| VGlut-F-400056 | 15933 | DvGlutMARCM-F329_seg1 | VGlut-Gal4 | F | 34455.66 | 0.77 |
| VGlut-F-000060 | 15986 | DvGlutMARCM-F376_seg1 | VGlut-Gal4 | F | 33946.75 | 0.74 |
| VGlut-F-000115 | 16110 | DvGlutMARCM-F493_seg1 | VGlut-Gal4 | F | 34696.60 | 0.77 |
| VGlut-F-000117 | 16121 | DvGlutMARCM-F504_seg1 | VGlut-Gal4 | F | 34205.51 | 0.75 |