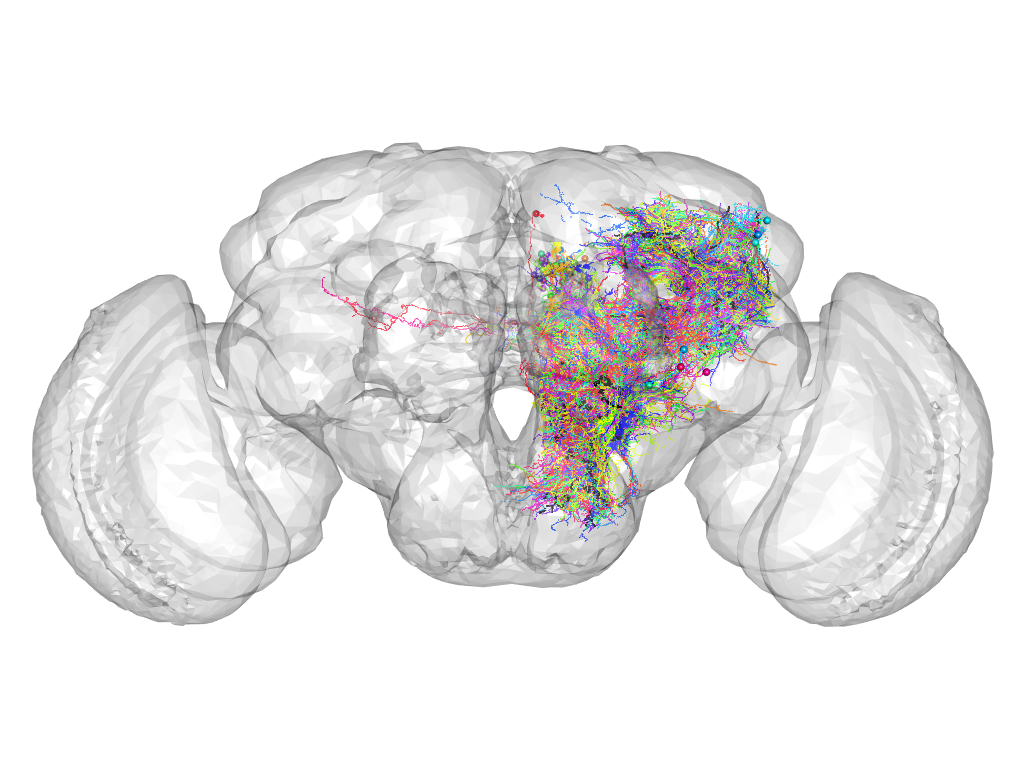
Cluster 872
Part of supercluster IX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-F-100101), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 666 | TH-F-200011 | IX | 0.70 |
| 873 | TH-F-000089 | IX | 0.51 |
| 406 | fru-F-000053 | VII | 0.45 |
| 861 | TH-F-000013 | IX | 0.28 |
| 484 | VGlut-F-000609 | IX | 0.27 |
Cluster composition
This cluster has 63 neurons. The exemplar of this cluster (TH-F-100101) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-F-100101 | 13738 | THMARCM-766F_seg1 | TH-Gal4 | F | 205085.86 | 1.00 |
| fru-M-600093 | 20 | FruMARCM-M002213_seg001 | fru-Gal4 | M | 140862.12 | 0.69 |
| fru-M-200246 | 882 | FruMARCM-M002147_seg001 | fru-Gal4 | M | 143164.39 | 0.69 |
| fru-M-100135 | 1221 | FruMARCM-M001405_seg001 | fru-Gal4 | M | 139093.31 | 0.70 |
| fru-M-100081 | 1519 | FruMARCM-M001064_seg002 | fru-Gal4 | M | 141293.70 | 0.68 |
| fru-M-400054 | 1685 | FruMARCM-M000839_seg001 | fru-Gal4 | M | 141228.12 | 0.69 |
| TH-M-100033 | 2584 | THMARCM-231M_seg1 | TH-Gal4 | M | 148598.24 | 0.73 |
| TH-M-100003 | 3089 | THMARCM-069M_seg1 | TH-Gal4 | M | 147399.29 | 0.72 |
| TH-M-100005 | 3091 | THMARCM-070M_seg1 | TH-Gal4 | M | 151236.09 | 0.71 |
| TH-M-100006 | 3092 | THMARCM-072M_seg1 | TH-Gal4 | M | 145477.93 | 0.71 |
| TH-M-000001 | 3096 | THMARCM-075M_seg1 | TH-Gal4 | M | 147127.93 | 0.73 |
| TH-M-000002 | 3097 | THMARCM-076M_seg1 | TH-Gal4 | M | 145894.59 | 0.73 |
| TH-M-100008 | 3100 | THMARCM-078M_seg2 | TH-Gal4 | M | 145069.78 | 0.68 |
| TH-M-100010 | 3102 | THMARCM-081M_seg1 | TH-Gal4 | M | 146897.98 | 0.69 |
| TH-M-100013 | 3120 | THMARCM-150M_seg2 | TH-Gal4 | M | 146756.22 | 0.72 |
| TH-M-400002 | 3127 | THMARCM-167M_seg1 | TH-Gal4 | M | 135682.09 | 0.70 |
| TH-M-300020 | 3129 | THMARCM-171M_seg1 | TH-Gal4 | M | 151490.46 | 0.75 |
| TH-M-300030 | 3216 | THMARCM-346M_seg1 | TH-Gal4 | M | 145880.72 | 0.71 |
| TH-M-300036 | 3230 | THMARCM-365M_seg1 | TH-Gal4 | M | 144831.82 | 0.71 |
| TH-M-200055 | 3236 | THMARCM-380M_seg1 | TH-Gal4 | M | 141904.72 | 0.70 |
| TH-M-200062 | 3244 | THMARCM-401M_seg1 | TH-Gal4 | M | 142658.39 | 0.71 |
| TH-M-000031 | 3331 | THMARCM-603M-X2_seg1 | TH-Gal4 | M | 140092.46 | 0.69 |
| TH-M-100083 | 3366 | THMARCM-779M_seg1 | TH-Gal4 | M | 145356.65 | 0.72 |
| TH-M-000063 | 3371 | THMARCM-802M_seg1 | TH-Gal4 | M | 136571.66 | 0.69 |
| TH-M-000064 | 3372 | THMARCM-803M_seg1 | TH-Gal4 | M | 139390.43 | 0.69 |
| TH-M-000065 | 3373 | THMARCM-804M_seg1 | TH-Gal4 | M | 147868.13 | 0.71 |
| TH-M-000069 | 3377 | THMARCM-808M_seg1 | TH-Gal4 | M | 143962.69 | 0.71 |
| fru-F-100083 | 3764 | FruMARCM-F002357_seg001 | fru-Gal4 | F | 143812.49 | 0.71 |
| Cha-F-000263 | 5193 | ChaMARCM-F001228_seg001 | Cha-Gal4 | F | 97930.59 | 0.62 |
| fru-F-700132 | 6294 | FruMARCM-F001392_seg001 | fru-Gal4 | F | 140202.91 | 0.69 |
| fru-F-400111 | 7315 | FruMARCM-F000954_seg001 | fru-Gal4 | F | 115722.42 | 0.66 |
| VGlut-F-800203 | 8887 | DvGlutMARCM-F003469_seg001 | VGlut-Gal4 | F | 138510.05 | 0.69 |
| Gad1-F-300122 | 9617 | GadMARCM-F000478_seg001 | Gad1-Gal4 | F | 49821.47 | 0.45 |
| Gad1-F-500085 | 9702 | GadMARCM-F000546_seg001 | Gad1-Gal4 | F | 53827.58 | 0.44 |
| fru-F-500029 | 9910 | FruMARCM-F000496_seg001 | fru-Gal4 | F | 131524.69 | 0.69 |
| VGlut-F-500479 | 10980 | DvGlutMARCM-F002870_seg001 | VGlut-Gal4 | F | 82868.01 | 0.56 |
| Gad1-F-400039 | 11376 | GadMARCM-F000329_seg001 | Gad1-Gal4 | F | 78042.07 | 0.54 |
| VGlut-F-400519 | 11808 | DvGlutMARCM-F002373_seg001 | VGlut-Gal4 | F | 145407.81 | 0.72 |
| TH-F-100003 | 13318 | THMARCM-036F_seg1 | TH-Gal4 | F | 144082.60 | 0.71 |
| TH-F-000005 | 13337 | THMARCM-068F_seg1 | TH-Gal4 | F | 143715.14 | 0.70 |
| TH-F-000007 | 13342 | THMARCM-095F_seg2 | TH-Gal4 | F | 146693.40 | 0.72 |
| TH-F-100020 | 13416 | THMARCM-223F_seg1 | TH-Gal4 | F | 148403.87 | 0.74 |
| TH-F-200021 | 13454 | THMARCM-292F_seg1 | TH-Gal4 | F | 138868.50 | 0.69 |
| TH-F-200035 | 13475 | THMARCM-314F_seg2 | TH-Gal4 | F | 148763.26 | 0.72 |
| TH-F-200036 | 13476 | THMARCM-323F_seg1 | TH-Gal4 | F | 147012.37 | 0.72 |
| TH-F-300048 | 13502 | THMARCM-376F_seg1 | TH-Gal4 | F | 151458.87 | 0.75 |
| TH-F-100061 | 13601 | THMARCM-538F_seg1 | TH-Gal4 | F | 146902.61 | 0.71 |
| TH-F-500002 | 13650 | THMARCM-657F_seg | TH-Gal4 | F | 143963.21 | 0.71 |
| TH-F-000066 | 13706 | THMARCM-734F_seg1 | TH-Gal4 | F | 151938.09 | 0.73 |
| TH-F-100097 | 13731 | THMARCM-760F_seg2 | TH-Gal4 | F | 149122.53 | 0.72 |
| TH-F-000081 | 13743 | THMARCM-776F_seg1 | TH-Gal4 | F | 150036.34 | 0.72 |
| TH-F-000084 | 13749 | THMARCM-786F_seg1 | TH-Gal4 | F | 148931.89 | 0.72 |
| TH-F-000086 | 13753 | THMARCM-790F_seg1 | TH-Gal4 | F | 150305.19 | 0.74 |
| TH-F-000096 | 13763 | THMARCM-814F_seg1 | TH-Gal4 | F | 145757.86 | 0.71 |
| TH-F-000098 | 13764 | THMARCM-816F_seg1 | TH-Gal4 | F | 147898.03 | 0.72 |
| TH-F-000102 | 13768 | THMARCM-820F_seg1 | TH-Gal4 | F | 149494.51 | 0.73 |
| TH-F-200129 | 13776 | THMARCM-828F_seg1 | TH-Gal4 | F | 142768.65 | 0.71 |
| Trh-F-300017 | 13907 | TPHMARCM-171F_seg1 | Trh-Gal4 | F | 76559.86 | 0.40 |
| VGlut-F-000200 | 14576 | DvGlutMARCM-F841_seg1 | VGlut-Gal4 | F | 146456.80 | 0.71 |
| VGlut-F-100128 | 14905 | DvGlutMARCM-F1248_seg1 | VGlut-Gal4 | F | 95272.09 | 0.61 |
| VGlut-F-100132 | 14947 | DvGlutMARCM-F1285_seg1 | VGlut-Gal4 | F | 143795.64 | 0.70 |
| VGlut-F-100147 | 15048 | DvGlutMARCM-F1387_seg1 | VGlut-Gal4 | F | 142757.70 | 0.70 |
| VGlut-F-400048 | 15894 | DvGlutMARCM-F295_seg1 | VGlut-Gal4 | F | 89752.45 | 0.46 |