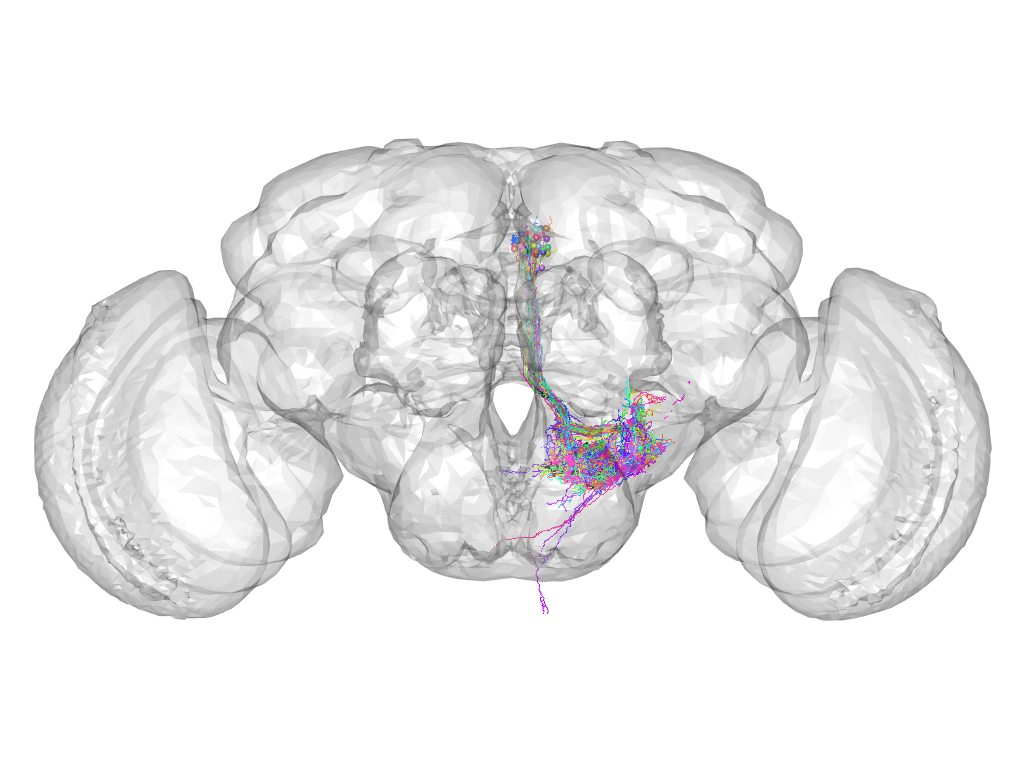
Cluster 441
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-600718), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 690 | VGlut-F-400061 | VIII | 0.79 |
| 430 | VGlut-F-500830 | VIII | 0.68 |
| 765 | VGlut-F-500530 | VIII | 0.64 |
| 652 | Tdc2-F-200010 | VIII | 0.62 |
| 691 | VGlut-F-700120 | VIII | 0.60 |
| 801 | Trh-F-600102 | VIII | 0.59 |
| 804 | Trh-F-500188 | VIII | 0.59 |
| 685 | VGlut-F-400542 | VIII | 0.58 |
| 751 | VGlut-F-400648 | VIII | 0.54 |
| 546 | VGlut-F-000492 | VIII | 0.52 |
Cluster composition
This cluster has 52 neurons. The exemplar of this cluster (VGlut-F-600718) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-600718 | 6790 | DvGlutMARCM-F003983_seg001 | VGlut-Gal4 | F | 20523.39 | 1.00 |
| VGlut-F-700560 | 5732 | DvGlutMARCM-F004439_seg002 | VGlut-Gal4 | F | 15431.29 | 0.76 |
| VGlut-F-600420 | 6043 | DvGlutMARCM-F002749_seg001 | VGlut-Gal4 | F | 15074.71 | 0.71 |
| VGlut-F-100379 | 6634 | DvGlutMARCM-F004383_seg001 | VGlut-Gal4 | F | 15410.44 | 0.73 |
| VGlut-F-600723 | 6837 | DvGlutMARCM-F004020_seg001 | VGlut-Gal4 | F | 15650.75 | 0.73 |
| VGlut-F-800256 | 6970 | DvGlutMARCM-F004117_seg001 | VGlut-Gal4 | F | 16338.76 | 0.78 |
| Cha-F-700048 | 8133 | ChaMARCM-F000175_seg001 | Cha-Gal4 | F | 15779.43 | 0.74 |
| VGlut-F-500538 | 8475 | DvGlutMARCM-F003071_seg001 | VGlut-Gal4 | F | 15653.59 | 0.77 |
| VGlut-F-700268 | 8501 | DvGlutMARCM-F003085_seg001 | VGlut-Gal4 | F | 16251.35 | 0.79 |
| VGlut-F-500709 | 8800 | DvGlutMARCM-F003611_seg004 | VGlut-Gal4 | F | 15314.27 | 0.77 |
| VGlut-F-600564 | 9177 | DvGlutMARCM-F003251_seg001 | VGlut-Gal4 | F | 14964.47 | 0.71 |
| VGlut-F-500617 | 9197 | DvGlutMARCM-F003264_seg002 | VGlut-Gal4 | F | 15888.55 | 0.78 |
| VGlut-F-500625 | 9206 | DvGlutMARCM-F003271_seg001 | VGlut-Gal4 | F | 14165.31 | 0.72 |
| VGlut-F-700328 | 9273 | DvGlutMARCM-F003328_seg003 | VGlut-Gal4 | F | 15085.63 | 0.76 |
| VGlut-F-300457 | 9311 | DvGlutMARCM-F003152_seg001 | VGlut-Gal4 | F | 15839.69 | 0.77 |
| VGlut-F-500605 | 9386 | DvGlutMARCM-F003215_seg001 | VGlut-Gal4 | F | 15047.85 | 0.72 |
| VGlut-F-600542 | 9393 | DvGlutMARCM-F003220_seg001 | VGlut-Gal4 | F | 15642.12 | 0.76 |
| VGlut-F-500346 | 10250 | DvGlutMARCM-F2083_seg1 | VGlut-Gal4 | F | 14899.43 | 0.72 |
| VGlut-F-700122 | 10848 | DvGlutMARCM-F002775_seg001 | VGlut-Gal4 | F | 15449.72 | 0.68 |
| VGlut-F-600436 | 10928 | DvGlutMARCM-F002824_seg001 | VGlut-Gal4 | F | 15160.61 | 0.74 |
| VGlut-F-600472 | 11116 | DvGlutMARCM-F002982_seg001 | VGlut-Gal4 | F | 15776.19 | 0.75 |
| VGlut-F-500381 | 11925 | DvGlutMARCM-F002459_seg001 | VGlut-Gal4 | F | 15163.83 | 0.74 |
| VGlut-F-600275 | 11988 | DvGlutMARCM-F002516_seg001 | VGlut-Gal4 | F | 14522.00 | 0.72 |
| VGlut-F-600365 | 12206 | DvGlutMARCM-F002673_seg002 | VGlut-Gal4 | F | 15663.59 | 0.77 |
| VGlut-F-600377 | 12223 | DvGlutMARCM-F002689_seg001 | VGlut-Gal4 | F | 15851.47 | 0.79 |
| VGlut-F-600387 | 12242 | DvGlutMARCM-F002704_seg001 | VGlut-Gal4 | F | 16038.15 | 0.74 |
| VGlut-F-600410 | 12278 | DvGlutMARCM-F002734_seg001 | VGlut-Gal4 | F | 15459.81 | 0.75 |
| VGlut-F-500302 | 12876 | DvGlutMARCM-F1917_seg1 | VGlut-Gal4 | F | 15634.05 | 0.75 |
| VGlut-F-400477 | 12951 | DvGlutMARCM-F1983_seg1 | VGlut-Gal4 | F | 14119.00 | 0.70 |
| VGlut-F-500353 | 13076 | DvGlutMARCM-F2099_seg1 | VGlut-Gal4 | F | 15522.28 | 0.73 |
| VGlut-F-400171 | 14433 | DvGlutMARCM-F724_seg1 | VGlut-Gal4 | F | 15313.01 | 0.69 |
| VGlut-F-400177 | 14443 | DvGlutMARCM-F734_seg1 | VGlut-Gal4 | F | 14797.20 | 0.71 |
| VGlut-F-400188 | 14455 | DvGlutMARCM-F742-x3_seg3 | VGlut-Gal4 | F | 15266.85 | 0.72 |
| VGlut-F-400221 | 14525 | DvGlutMARCM-F796_seg1 | VGlut-Gal4 | F | 15857.65 | 0.72 |
| VGlut-F-400248 | 14594 | DvGlutMARCM-F858_seg1 | VGlut-Gal4 | F | 15698.00 | 0.77 |
| VGlut-F-400281 | 14682 | DvGlutMARCM-F942_seg1 | VGlut-Gal4 | F | 15659.71 | 0.74 |
| VGlut-F-500141 | 14790 | DvGlutMARCM-F1141_seg1 | VGlut-Gal4 | F | 15540.34 | 0.73 |
| VGlut-F-500142 | 14791 | DvGlutMARCM-F1142_seg1 | VGlut-Gal4 | F | 15759.63 | 0.75 |
| VGlut-F-500144 | 14793 | DvGlutMARCM-F1143_seg1 | VGlut-Gal4 | F | 15754.29 | 0.69 |
| VGlut-F-500179 | 15176 | DvGlutMARCM-F1506_seg2 | VGlut-Gal4 | F | 15752.66 | 0.69 |
| VGlut-F-500190 | 15225 | DvGlutMARCM-F1549_seg1 | VGlut-Gal4 | F | 15173.13 | 0.64 |
| VGlut-F-500191 | 15226 | DvGlutMARCM-F1549_seg2 | VGlut-Gal4 | F | 15618.56 | 0.65 |
| VGlut-F-500223 | 15328 | DvGlutMARCM-F1669_seg1 | VGlut-Gal4 | F | 15461.63 | 0.76 |
| VGlut-F-600086 | 15346 | DvGlutMARCM-F1695_seg1 | VGlut-Gal4 | F | 15334.57 | 0.76 |
| VGlut-F-600092 | 15400 | DvGlutMARCM-F1761_seg1 | VGlut-Gal4 | F | 14446.98 | 0.65 |
| VGlut-F-500262 | 15431 | DvGlutMARCM-F1788_seg1 | VGlut-Gal4 | F | 16147.74 | 0.73 |
| VGlut-F-500106 | 15490 | DvGlutMARCM-F647_seg1 | VGlut-Gal4 | F | 15967.41 | 0.69 |
| VGlut-F-400117 | 15503 | DvGlutMARCM-F656-X2_seg1 | VGlut-Gal4 | F | 16214.99 | 0.78 |
| VGlut-F-400025 | 15715 | DvGlutMARCM-F127-X3_seg2 | VGlut-Gal4 | F | 16017.62 | 0.70 |
| VGlut-F-500045 | 15845 | DvGlutMARCM-F252_seg1 | VGlut-Gal4 | F | 15502.04 | 0.74 |
| VGlut-F-500061 | 16023 | DvGlutMARCM-F413_seg1 | VGlut-Gal4 | F | 15079.77 | 0.74 |
| VGlut-F-500068 | 16031 | DvGlutMARCM-F422_seg1 | VGlut-Gal4 | F | 15914.68 | 0.78 |