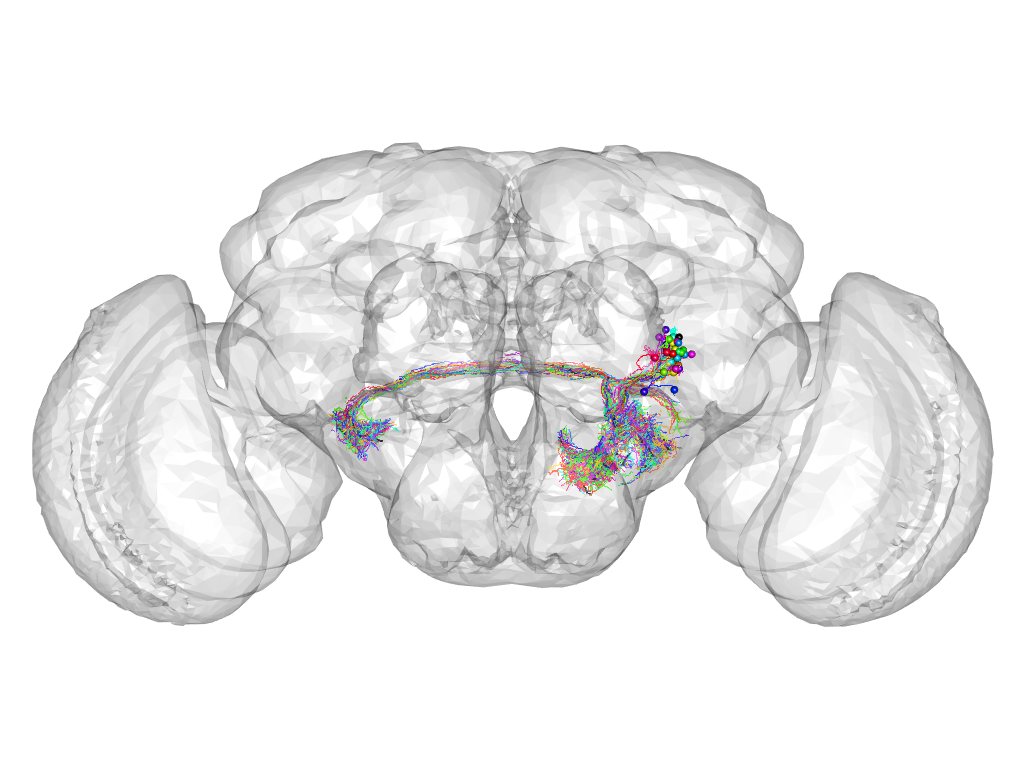
Cluster 801
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-600102), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 804 | Trh-F-500188 | VIII | 0.73 |
| 803 | Trh-F-700063 | VIII | 0.68 |
| 45 | fru-M-600070 | VIII | 0.48 |
| 652 | Tdc2-F-200010 | VIII | 0.47 |
| 960 | VGlut-F-500219 | VIII | 0.41 |
Cluster composition
This cluster has 37 neurons. The exemplar of this cluster (Trh-F-600102) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-600102 | 12463 | TPHMARCM-1252F_seg1 | Trh-Gal4 | F | 15079.34 | 1.00 |
| fru-M-000108 | 589 | FruMARCM-M001784_seg001 | fru-Gal4 | M | 10848.87 | 0.72 |
| Trh-M-000136 | 1728 | TPHMARCM-M001804_seg001 | Trh-Gal4 | M | 11071.00 | 0.70 |
| Trh-M-900033 | 2040 | TPHMARCM-M001550_seg001 | Trh-Gal4 | M | 11079.41 | 0.71 |
| Trh-M-600089 | 2142 | TPHMARCM-M001632_seg001 | Trh-Gal4 | M | 11392.50 | 0.74 |
| Trh-M-600091 | 2144 | TPHMARCM-M001633_seg002 | Trh-Gal4 | M | 11582.09 | 0.75 |
| Trh-M-600067 | 2610 | TPHMARCM-1316M_seg1 | Trh-Gal4 | M | 10793.56 | 0.72 |
| Trh-M-600071 | 2623 | TPHMARCM-M001357_seg001 | Trh-Gal4 | M | 11332.02 | 0.75 |
| Trh-M-600074 | 2626 | TPHMARCM-M001364_seg001 | Trh-Gal4 | M | 10967.83 | 0.74 |
| Trh-M-500149 | 2754 | TPHMARCM-M001521_seg001 | Trh-Gal4 | M | 10678.88 | 0.72 |
| Trh-M-100014 | 3395 | TPHMARCM-080M_seg1 | Trh-Gal4 | M | 10677.19 | 0.70 |
| Trh-M-100026 | 3418 | TPHMARCM-103M_seg1 | Trh-Gal4 | M | 10861.48 | 0.71 |
| Trh-M-600002 | 3543 | TPHMARCM-454M_seg1 | Trh-Gal4 | M | 11186.53 | 0.73 |
| Trh-M-600010 | 3588 | TPHMARCM-533M_seg1 | Trh-Gal4 | M | 11171.62 | 0.74 |
| Trh-M-500032 | 3607 | TPHMARCM-550M_seg1 | Trh-Gal4 | M | 11309.11 | 0.72 |
| Trh-M-600018 | 3608 | TPHMARCM-560M_seg1 | Trh-Gal4 | M | 10786.01 | 0.72 |
| Trh-M-500058 | 3688 | TPHMARCM-804M_seg1 | Trh-Gal4 | M | 11193.01 | 0.74 |
| Trh-F-600074 | 12400 | TPHMARCM-1197F_seg1 | Trh-Gal4 | F | 11448.54 | 0.75 |
| Trh-F-700058 | 12413 | TPHMARCM-1206F_seg1 | Trh-Gal4 | F | 11248.87 | 0.73 |
| Trh-F-600085 | 12434 | TPHMARCM-1229F_seg1 | Trh-Gal4 | F | 11404.68 | 0.75 |
| Trh-F-600088 | 12440 | TPHMARCM-1234F_seg1 | Trh-Gal4 | F | 10985.29 | 0.73 |
| Trh-F-600093 | 12445 | TPHMARCM-1239F_seg1 | Trh-Gal4 | F | 11270.81 | 0.75 |
| Trh-F-700064 | 12483 | TPHMARCM-1272F_seg1 | Trh-Gal4 | F | 11350.97 | 0.73 |
| Trh-F-500150 | 12487 | TPHMARCM-1275F_seg1 | Trh-Gal4 | F | 10867.30 | 0.73 |
| Trh-F-400081 | 12718 | TPHMARCM-880F_seg1 | Trh-Gal4 | F | 10749.23 | 0.72 |
| Trh-F-600037 | 13224 | TPHMARCM-709F_seg1 | Trh-Gal4 | F | 11260.49 | 0.74 |
| Trh-F-900014 | 13257 | TPHMARCM-746F_seg1 | Trh-Gal4 | F | 10888.35 | 0.72 |
| Trh-F-900020 | 13277 | TPHMARCM-763F_seg1 | Trh-Gal4 | F | 11423.22 | 0.74 |
| Trh-F-600053 | 13297 | TPHMARCM-780F_seg1 | Trh-Gal4 | F | 11135.25 | 0.73 |
| Trh-F-400006 | 13833 | TPHMARCM-054F_seg1 | Trh-Gal4 | F | 10480.06 | 0.72 |
| Trh-F-900002 | 13921 | TPHMARCM-209F_seg1 | Trh-Gal4 | F | 10728.28 | 0.71 |
| Trh-F-200024 | 13952 | TPHMARCM-243F_seg1 | Trh-Gal4 | F | 10054.32 | 0.67 |
| Trh-F-500051 | 14156 | TPHMARCM-580F_seg1 | Trh-Gal4 | F | 11201.99 | 0.72 |
| Trh-F-300082 | 14167 | TPHMARCM-594F_seg1 | Trh-Gal4 | F | 11428.73 | 0.75 |
| Trh-F-600035 | 14209 | TPHMARCM-649F_seg2 | Trh-Gal4 | F | 11418.88 | 0.72 |
| Trh-F-500078 | 14241 | TPHMARCM-694F_seg2 | Trh-Gal4 | F | 10725.36 | 0.71 |
| VGlut-F-600077 | 15210 | DvGlutMARCM-F1538_seg1 | VGlut-Gal4 | F | 10936.17 | 0.73 |