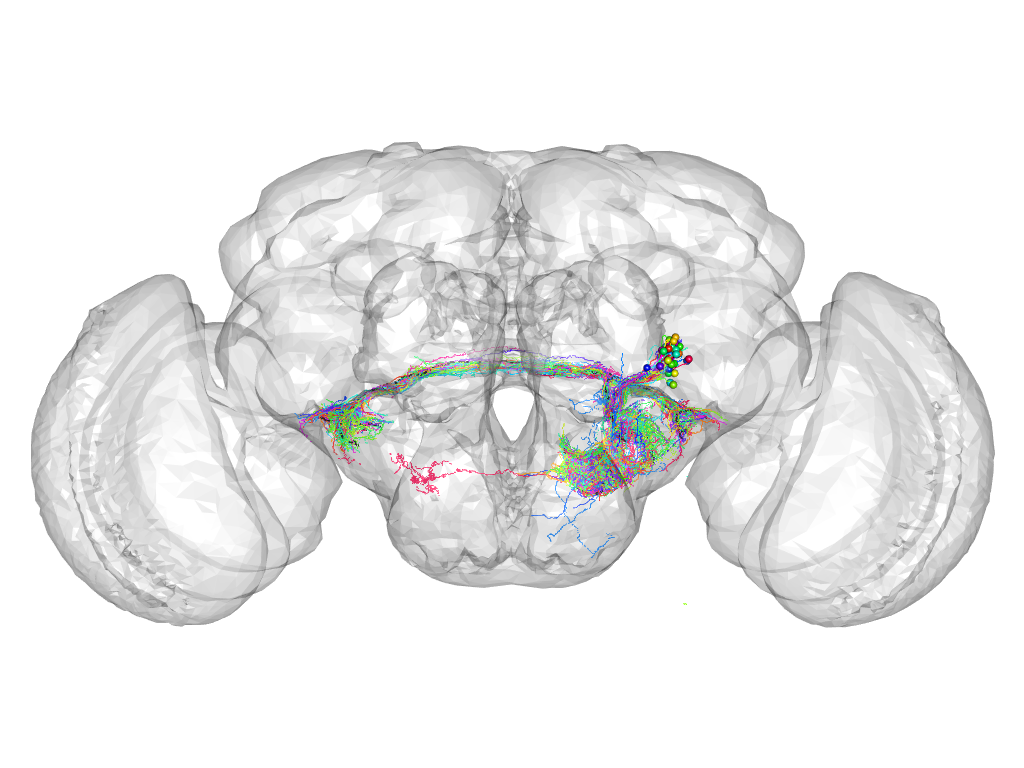
Cluster 804
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-500188), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 801 | Trh-F-600102 | VIII | 0.69 |
| 803 | Trh-F-700063 | VIII | 0.57 |
| 45 | fru-M-600070 | VIII | 0.55 |
| 652 | Tdc2-F-200010 | VIII | 0.45 |
| 960 | VGlut-F-500219 | VIII | 0.42 |
Cluster composition
This cluster has 42 neurons. The exemplar of this cluster (Trh-F-500188) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-500188 | 12546 | TPHMARCM-1334F_seg1 | Trh-Gal4 | F | 13951.81 | 1.00 |
| fru-M-200297 | 262 | FruMARCM-M002488_seg001 | fru-Gal4 | M | 10013.09 | 0.71 |
| fru-M-700093 | 474 | FruMARCM-M001577_seg001 | fru-Gal4 | M | 9801.60 | 0.71 |
| fru-M-800069 | 610 | FruMARCM-M001799_seg001 | fru-Gal4 | M | 9641.34 | 0.69 |
| fru-M-000130 | 745 | FruMARCM-M001959_seg001 | fru-Gal4 | M | 9263.34 | 0.68 |
| fru-M-700079 | 1211 | FruMARCM-M001389_seg001 | fru-Gal4 | M | 9297.32 | 0.66 |
| fru-M-100085 | 1523 | FruMARCM-M001066_seg001 | fru-Gal4 | M | 10348.09 | 0.73 |
| fru-M-100090 | 1528 | FruMARCM-M001069_seg001 | fru-Gal4 | M | 10250.79 | 0.74 |
| fru-M-200080 | 1600 | FruMARCM-M000695_seg001 | fru-Gal4 | M | 10057.22 | 0.73 |
| Trh-M-900048 | 2058 | TPHMARCM-M001563_seg001 | Trh-Gal4 | M | 10253.87 | 0.73 |
| fru-M-600007 | 2415 | FruMARCM-M000136_seg001 | fru-Gal4 | M | 9634.46 | 0.69 |
| Trh-M-200001 | 2577 | TPHMARCM-105M_seg1 | Trh-Gal4 | M | 9689.74 | 0.70 |
| Trh-M-600073 | 2625 | TPHMARCM-M001359_seg001 | Trh-Gal4 | M | 9236.34 | 0.65 |
| Trh-M-400070 | 2841 | TPHMARCM-1067M_seg1 | Trh-Gal4 | M | 10471.89 | 0.72 |
| Trh-M-300099 | 2854 | TPHMARCM-1082M_seg2 | Trh-Gal4 | M | 10092.46 | 0.71 |
| Trh-M-700013 | 2979 | TPHMARCM-918M_seg1 | Trh-Gal4 | M | 10460.70 | 0.74 |
| Trh-M-100056 | 2987 | TPHMARCM-925M_seg1 | Trh-Gal4 | M | 10204.97 | 0.73 |
| Trh-M-800016 | 2989 | TPHMARCM-931M_seg1 | Trh-Gal4 | M | 10025.44 | 0.72 |
| Trh-M-400048 | 3018 | TPHMARCM-970M_seg1 | Trh-Gal4 | M | 10415.16 | 0.74 |
| Trh-M-500027 | 3384 | TPHMARCM-502M_seg1 | Trh-Gal4 | M | 10037.03 | 0.71 |
| Trh-M-000009 | 3405 | TPHMARCM-091M_seg1 | Trh-Gal4 | M | 10551.37 | 0.73 |
| Trh-M-400005 | 3497 | TPHMARCM-367M_seg1 | Trh-Gal4 | M | 9992.05 | 0.72 |
| Trh-M-600001 | 3538 | TPHMARCM-444M_seg1 | Trh-Gal4 | M | 10164.03 | 0.73 |
| VGlut-F-700454 | 6733 | DvGlutMARCM-F003940_seg001 | VGlut-Gal4 | F | 9769.57 | 0.68 |
| VGlut-F-000620 | 6835 | DvGlutMARCM-F004018_seg001 | VGlut-Gal4 | F | 9401.68 | 0.69 |
| Cha-F-100060 | 7085 | ChaMARCM-F000565_seg001 | Cha-Gal4 | F | 8317.25 | 0.49 |
| VGlut-F-500710 | 8801 | DvGlutMARCM-F003612_seg001 | VGlut-Gal4 | F | 9241.92 | 0.67 |
| VGlut-F-700247 | 11823 | DvGlutMARCM-F002386_seg002 | VGlut-Gal4 | F | 9565.82 | 0.62 |
| Trh-F-600082 | 12423 | TPHMARCM-1214F_seg1 | Trh-Gal4 | F | 9110.64 | 0.68 |
| Trh-F-600083 | 12432 | TPHMARCM-1228F_seg1 | Trh-Gal4 | F | 10438.63 | 0.74 |
| Trh-F-700026 | 13266 | TPHMARCM-753F_seg2 | Trh-Gal4 | F | 9310.08 | 0.70 |
| Trh-F-900022 | 13298 | TPHMARCM-781F_seg1 | Trh-Gal4 | F | 10324.95 | 0.73 |
| Trh-F-500010 | 13857 | TPHMARCM-127F_seg1 | Trh-Gal4 | F | 10363.84 | 0.74 |
| Trh-F-300006 | 13885 | TPHMARCM-150F_seg1 | Trh-Gal4 | F | 10195.30 | 0.73 |
| Trh-F-000018 | 13916 | TPHMARCM-199F_seg1 | Trh-Gal4 | F | 10190.05 | 0.74 |
| Trh-F-000020 | 13929 | TPHMARCM-219F_seg1 | Trh-Gal4 | F | 8802.93 | 0.64 |
| Trh-F-500057 | 14192 | TPHMARCM-634F-01_seg1 | Trh-Gal4 | F | 9913.96 | 0.72 |
| Trh-F-500071 | 14234 | TPHMARCM-686F_seg1 | Trh-Gal4 | F | 10255.81 | 0.73 |
| Trh-F-900030 | 14298 | TPHMARCM-838F_seg1 | Trh-Gal4 | F | 5653.12 | 0.51 |
| VGlut-F-500137 | 14660 | DvGlutMARCM-F921_seg1 | VGlut-Gal4 | F | 10358.90 | 0.73 |
| VGlut-F-900010 | 14886 | DvGlutMARCM-F1230_seg1 | VGlut-Gal4 | F | 7617.12 | 0.38 |
| VGlut-F-500182 | 15214 | DvGlutMARCM-F1540_seg1 | VGlut-Gal4 | F | 10241.31 | 0.74 |