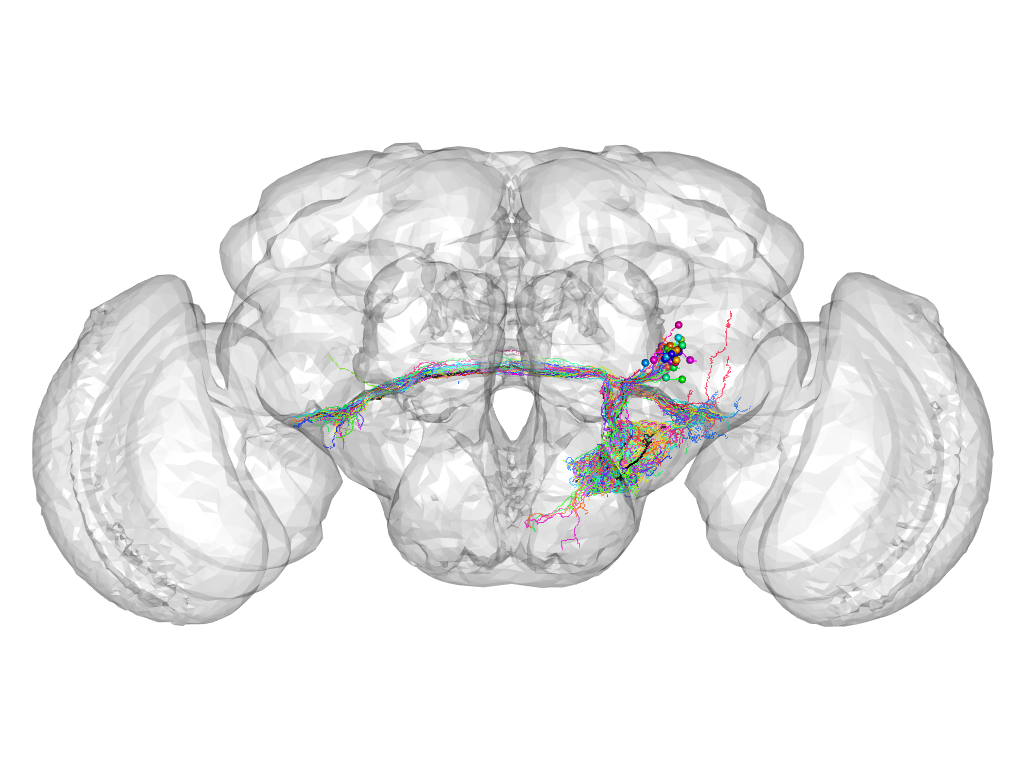
Cluster 45
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-600070), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 804 | Trh-F-500188 | VIII | 0.63 |
| 801 | Trh-F-600102 | VIII | 0.53 |
| 803 | Trh-F-700063 | VIII | 0.41 |
| 960 | VGlut-F-500219 | VIII | 0.38 |
| 652 | Tdc2-F-200010 | VIII | 0.37 |
Cluster composition
This cluster has 36 neurons. The exemplar of this cluster (fru-M-600070) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-600070 | 494 | FruMARCM-M001591_seg001 | fru-Gal4 | M | 13940.42 | 1.00 |
| fru-M-200325 | 8 | FruMARCM-M002578_seg002 | fru-Gal4 | M | 9556.10 | 0.69 |
| fru-M-600097 | 72 | FruMARCM-M002270_seg001 | fru-Gal4 | M | 10421.21 | 0.73 |
| fru-M-500233 | 166 | FruMARCM-M002386_seg001 | fru-Gal4 | M | 10154.30 | 0.67 |
| fru-M-200300 | 267 | FruMARCM-M002493_seg001 | fru-Gal4 | M | 10766.76 | 0.75 |
| fru-M-700184 | 286 | FruMARCM-M002510_seg003 | fru-Gal4 | M | 9879.52 | 0.68 |
| fru-M-300183 | 454 | FruMARCM-M001563_seg001 | fru-Gal4 | M | 9186.65 | 0.68 |
| fru-M-100158 | 528 | FruMARCM-M001649_seg001 | fru-Gal4 | M | 9617.61 | 0.68 |
| fru-M-800070 | 611 | FruMARCM-M001800_seg001 | fru-Gal4 | M | 10059.77 | 0.68 |
| fru-M-200220 | 640 | FruMARCM-M001849_seg001 | fru-Gal4 | M | 9592.71 | 0.67 |
| fru-M-500176 | 684 | FruMARCM-M001888_seg001 | fru-Gal4 | M | 9015.68 | 0.67 |
| fru-M-500183 | 704 | FruMARCM-M001906_seg001 | fru-Gal4 | M | 10015.05 | 0.71 |
| fru-M-400187 | 810 | FruMARCM-M002018_seg001 | fru-Gal4 | M | 10409.72 | 0.72 |
| fru-M-500213 | 855 | FruMARCM-M002108_seg001 | fru-Gal4 | M | 10055.12 | 0.70 |
| fru-M-800044 | 985 | FruMARCM-M001165_seg001 | fru-Gal4 | M | 8606.41 | 0.58 |
| fru-M-500114 | 1026 | FruMARCM-M001206_seg001 | fru-Gal4 | M | 10026.70 | 0.70 |
| fru-M-300138 | 1070 | FruMARCM-M001248_seg001 | fru-Gal4 | M | 9354.43 | 0.65 |
| fru-M-300152 | 1161 | FruMARCM-M001348_seg002 | fru-Gal4 | M | 7760.18 | 0.62 |
| fru-M-900033 | 1179 | FruMARCM-M001362_seg001 | fru-Gal4 | M | 10407.65 | 0.72 |
| fru-M-600069 | 1276 | FruMARCM-M001463_seg001 | fru-Gal4 | M | 10096.89 | 0.70 |
| fru-M-100075 | 1497 | FruMARCM-M001049_seg001 | fru-Gal4 | M | 10051.26 | 0.71 |
| fru-M-600023 | 1855 | FruMARCM-M000372_seg001 | fru-Gal4 | M | 8466.76 | 0.63 |
| fru-M-600001 | 2409 | FruMARCM-M000130_seg001 | fru-Gal4 | M | 9417.38 | 0.61 |
| fru-M-900018 | 2437 | FruMARCM-M000154_seg001 | fru-Gal4 | M | 9335.35 | 0.68 |
| fru-M-500000 | 2475 | FruMARCM-M000231_seg001 | fru-Gal4 | M | 9622.12 | 0.65 |
| fru-F-600122 | 3704 | FruMARCM-F002192_seg001 | fru-Gal4 | F | 9525.69 | 0.71 |
| fru-F-100093 | 3829 | FruMARCM-F002653_seg001 | fru-Gal4 | F | 10202.12 | 0.72 |
| fru-F-300063 | 6195 | FruMARCM-F001124_seg001 | fru-Gal4 | F | 9639.55 | 0.68 |
| fru-F-100049 | 6278 | FruMARCM-F001308_seg001 | fru-Gal4 | F | 9991.35 | 0.71 |
| fru-F-600078 | 6307 | FruMARCM-F001426_seg001 | fru-Gal4 | F | 9824.06 | 0.70 |
| fru-F-700140 | 6311 | FruMARCM-F001430_seg001 | fru-Gal4 | F | 9661.09 | 0.70 |
| fru-F-700146 | 6335 | FruMARCM-F001485_seg001 | fru-Gal4 | F | 10030.42 | 0.65 |
| fru-F-700072 | 7279 | FruMARCM-F000913_seg001 | fru-Gal4 | F | 9354.44 | 0.64 |
| fru-F-700095 | 7330 | FruMARCM-F000967_seg001 | fru-Gal4 | F | 9062.97 | 0.64 |
| fru-F-500137 | 7821 | FruMARCM-F000714_seg001 | fru-Gal4 | F | 9380.07 | 0.66 |
| Cha-F-700012 | 10097 | ChaMARCM-F000053_seg002 | Cha-Gal4 | F | 3626.39 | 0.33 |