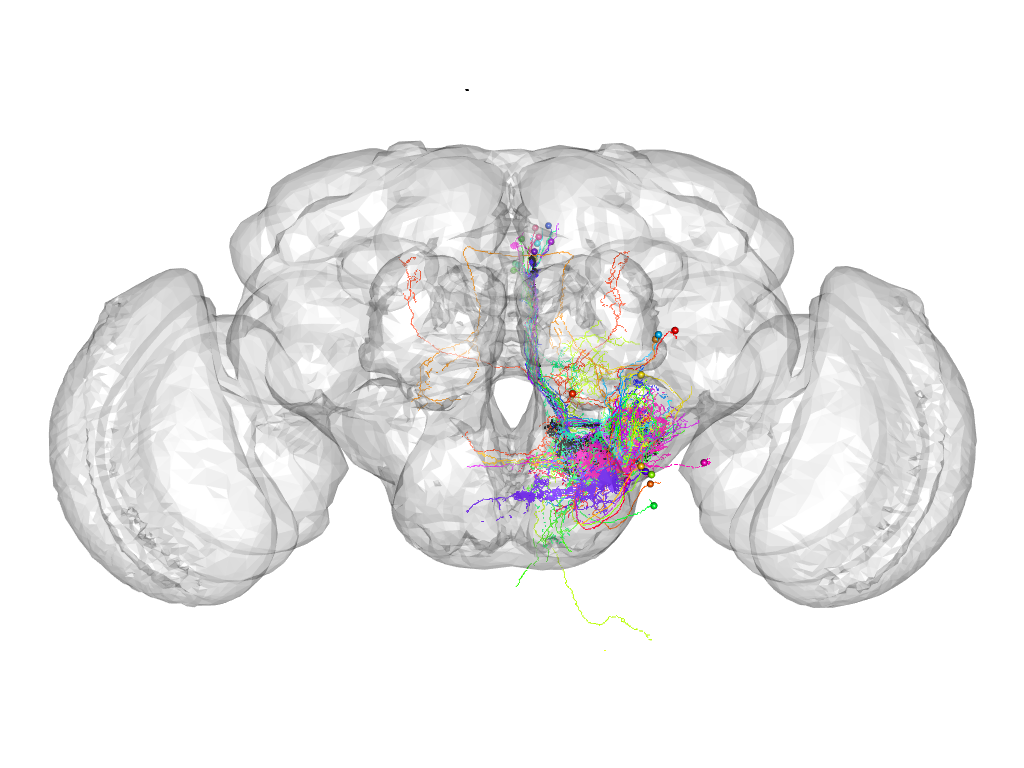
Cluster 690
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-400061), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 430 | VGlut-F-500830 | VIII | 0.63 |
| 652 | Tdc2-F-200010 | VIII | 0.62 |
| 801 | Trh-F-600102 | VIII | 0.56 |
| 751 | VGlut-F-400648 | VIII | 0.55 |
| 804 | Trh-F-500188 | VIII | 0.54 |
| 691 | VGlut-F-700120 | VIII | 0.53 |
| 441 | VGlut-F-600718 | VIII | 0.53 |
| 685 | VGlut-F-400542 | VIII | 0.51 |
Cluster composition
This cluster has 34 neurons. The exemplar of this cluster (VGlut-F-400061) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-400061 | 10824 | DvGlutMARCM-F338_seg1 | VGlut-Gal4 | F | 70077.93 | 1.00 |
| fru-M-300044 | 1821 | FruMARCM-M000341_seg002 | fru-Gal4 | M | 28778.02 | 0.47 |
| 5-HT1B-M-400006 | 2506 | 5HT1bMARCM-M000050_seg002 | 5-HT1B-Gal4 | M | 26080.32 | 0.19 |
| Cha-F-200190 | 4857 | ChaMARCM-F000979_seg001 | Cha-Gal4 | F | 38744.19 | 0.48 |
| Cha-F-300292 | 5087 | ChaMARCM-F001149_seg003 | Cha-Gal4 | F | 39278.86 | 0.43 |
| Cha-F-800061 | 5176 | ChaMARCM-F001215_seg001 | Cha-Gal4 | F | 19959.76 | 0.40 |
| Cha-F-600147 | 5689 | ChaMARCM-F000947_seg001 | Cha-Gal4 | F | 35054.26 | 0.55 |
| VGlut-F-500176 | 8428 | DvGlutMARCM-F1504_seg1 | VGlut-Gal4 | F | 43331.61 | 0.64 |
| VGlut-F-000135 | 10254 | DvGlutMARCM-F529_seg1 | VGlut-Gal4 | F | 36952.67 | 0.32 |
| VGlut-F-500035 | 10765 | DvGlutMARCM-F-0323-04-F204_seg1 | VGlut-Gal4 | F | 19169.49 | 0.40 |
| VGlut-F-700186 | 10910 | DvGlutMARCM-F002811_seg001 | VGlut-Gal4 | F | 48176.03 | 0.73 |
| VGlut-F-500272 | 12790 | DvGlutMARCM-F1838_seg1 | VGlut-Gal4 | F | 42345.07 | 0.47 |
| VGlut-F-200351 | 12958 | DvGlutMARCM-F1989_seg1 | VGlut-Gal4 | F | 41503.81 | 0.64 |
| VGlut-F-400305 | 14386 | DvGlutMARCM-F1066_seg1 | VGlut-Gal4 | F | 44756.48 | 0.59 |
| VGlut-F-400185 | 14452 | DvGlutMARCM-F741_seg1 | VGlut-Gal4 | F | 41790.96 | 0.53 |
| VGlut-F-600028 | 14480 | DvGlutMARCM-F759-x3_seg1 | VGlut-Gal4 | F | 47344.55 | 0.63 |
| VGlut-F-500108 | 14522 | DvGlutMARCM-F794_seg1 | VGlut-Gal4 | F | 36952.92 | 0.62 |
| VGlut-F-400233 | 14547 | DvGlutMARCM-F815_seg1 | VGlut-Gal4 | F | 44717.00 | 0.71 |
| VGlut-F-400266 | 14640 | DvGlutMARCM-F896_seg1 | VGlut-Gal4 | F | 9484.99 | 0.35 |
| VGlut-F-400274 | 14663 | DvGlutMARCM-F924_seg1 | VGlut-Gal4 | F | 30899.99 | 0.52 |
| VGlut-F-200186 | 14684 | DvGlutMARCM-F944_seg1 | VGlut-Gal4 | F | 45307.98 | 0.67 |
| VGlut-F-500234 | 15354 | DvGlutMARCM-F1703_seg1 | VGlut-Gal4 | F | 43632.89 | 0.67 |
| VGlut-F-500239 | 15363 | DvGlutMARCM-F1715_seg2 | VGlut-Gal4 | F | 44363.90 | 0.68 |
| VGlut-F-400135 | 15528 | DvGlutMARCM-F675-X2_seg2 | VGlut-Gal4 | F | 47649.41 | 0.67 |
| VGlut-F-400136 | 15529 | DvGlutMARCM-F676-X2_seg1 | VGlut-Gal4 | F | 49709.59 | 0.75 |
| VGlut-F-800010 | 15771 | DvGlutMARCM-F178_seg1 | VGlut-Gal4 | F | 50661.65 | 0.59 |
| VGlut-F-500033 | 15774 | DvGlutMARCM-F180_seg1 | VGlut-Gal4 | F | 40585.41 | 0.61 |
| VGlut-F-500034 | 15792 | DvGlutMARCM-F203_seg1 | VGlut-Gal4 | F | 44656.44 | 0.68 |
| VGlut-F-400042 | 15838 | DvGlutMARCM-F244_seg1 | VGlut-Gal4 | F | 33205.80 | 0.49 |
| VGlut-F-400057 | 15934 | DvGlutMARCM-F330_seg1 | VGlut-Gal4 | F | 46814.94 | 0.73 |
| VGlut-F-400073 | 15966 | DvGlutMARCM-F357_seg1 | VGlut-Gal4 | F | 42075.39 | 0.61 |
| VGlut-F-500065 | 16027 | DvGlutMARCM-F418_seg1 | VGlut-Gal4 | F | 41160.80 | 0.64 |
| VGlut-F-500067 | 16030 | DvGlutMARCM-F421_seg1 | VGlut-Gal4 | F | 47079.03 | 0.70 |
| VGlut-F-100038 | 16045 | DvGlutMARCM-F436_seg1 | VGlut-Gal4 | F | 46200.12 | 0.59 |