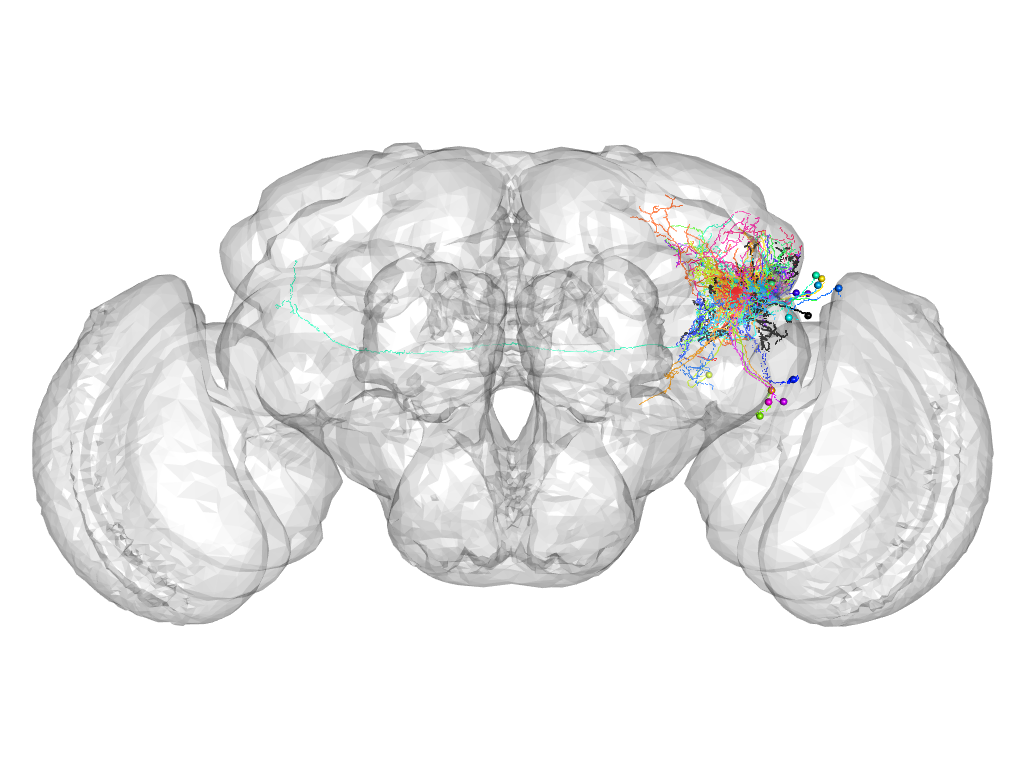
Cluster 674
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Gad1-F-600005), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 998 | VGlut-F-100009 | X | 0.65 |
| 621 | Gad1-F-500089 | XVII | 0.65 |
| 136 | TH-M-000013 | XVII | 0.64 |
| 662 | Tdc2-F-400026 | XIII | 0.63 |
| 872 | TH-F-100101 | IX | 0.57 |
| 52 | fru-M-000094 | X | 0.54 |
| 2 | fru-M-200253 | XVII | 0.52 |
| 440 | VGlut-F-600716 | XVII | 0.50 |
Cluster composition
This cluster has 21 neurons. The exemplar of this cluster (Gad1-F-600005) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Gad1-F-600005 | 10579 | GadMARCM-F000098_seg002 | Gad1-Gal4 | F | 54645.52 | 1.00 |
| Gad1-F-600088 | 3945 | GadMARCM-F000575_seg002 | Gad1-Gal4 | F | 25808.50 | 0.51 |
| Gad1-F-100074 | 4055 | GadMARCM-F000654_seg001 | Gad1-Gal4 | F | 32952.49 | 0.54 |
| Cha-F-200351 | 4343 | ChaMARCM-F001400_seg002 | Cha-Gal4 | F | 28519.91 | 0.53 |
| fru-F-400219 | 4800 | FruMARCM-F002134_seg001 | fru-Gal4 | F | 33752.93 | 0.66 |
| Cha-F-400222 | 4837 | ChaMARCM-F000963_seg001 | Cha-Gal4 | F | 29443.69 | 0.56 |
| Cha-F-200201 | 4915 | ChaMARCM-F001026_seg001 | Cha-Gal4 | F | 36513.18 | 0.66 |
| VGlut-F-500445 | 5970 | DvGlutMARCM-F002665_seg001 | VGlut-Gal4 | F | 30867.94 | 0.57 |
| fru-F-500177 | 6274 | FruMARCM-F001305_seg002 | fru-Gal4 | F | 28705.52 | 0.59 |
| VGlut-F-600752 | 6930 | DvGlutMARCM-F004085_seg001 | VGlut-Gal4 | F | 30098.65 | 0.61 |
| Cha-F-500135 | 7200 | ChaMARCM-F000644_seg003 | Cha-Gal4 | F | 25191.31 | 0.23 |
| Cha-F-300136 | 7675 | ChaMARCM-F000501_seg001 | Cha-Gal4 | F | 34510.22 | 0.68 |
| Cha-F-200034 | 8151 | ChaMARCM-F000182_seg003 | Cha-Gal4 | F | 35716.48 | 0.70 |
| Gad1-F-700003 | 10610 | GadMARCM-F000118_seg001 | Gad1-Gal4 | F | 33594.82 | 0.60 |
| VGlut-F-700213 | 10839 | DvGlutMARCM-F002839_seg002 | VGlut-Gal4 | F | 31704.60 | 0.57 |
| fru-F-400011 | 11468 | FruMARCM-F000080_seg002 | fru-Gal4 | F | 28351.49 | 0.60 |
| VGlut-F-500442 | 12187 | DvGlutMARCM-F002662_seg003 | VGlut-Gal4 | F | 29187.56 | 0.52 |
| VGlut-F-200257 | 14957 | DvGlutMARCM-F1295_seg1 | VGlut-Gal4 | F | 37419.43 | 0.71 |
| VGlut-F-600059 | 15067 | DvGlutMARCM-F1404_seg1 | VGlut-Gal4 | F | 31739.25 | 0.63 |
| VGlut-F-000390 | 15288 | DvGlutMARCM-F1619_seg1 | VGlut-Gal4 | F | 23867.81 | 0.32 |
| VGlut-F-600090 | 15375 | DvGlutMARCM-F1728_seg1 | VGlut-Gal4 | F | 30879.93 | 0.62 |