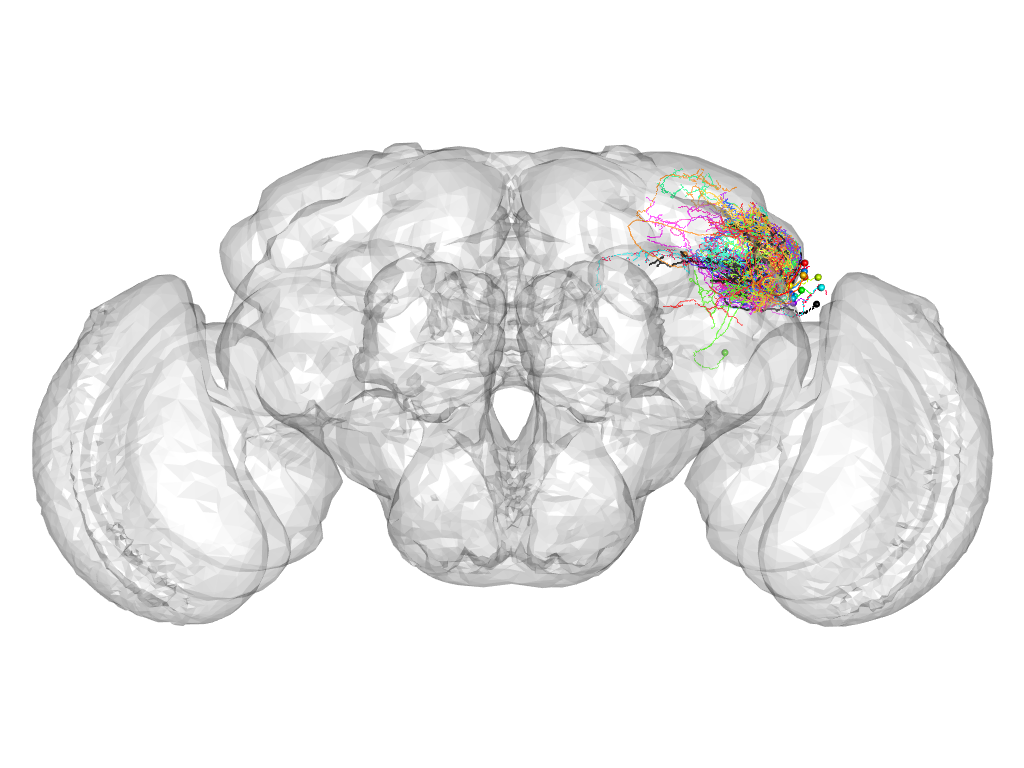
Cluster 2
Part of supercluster XVII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-200253), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 339 | Cha-F-100223 | XVII | 0.71 |
| 662 | Tdc2-F-400026 | XIII | 0.71 |
| 136 | TH-M-000013 | XVII | 0.67 |
| 674 | Gad1-F-600005 | X | 0.54 |
| 439 | VGlut-F-700465 | XI | 0.52 |
Cluster composition
This cluster has 25 neurons. The exemplar of this cluster (fru-M-200253) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-200253 | 18 | FruMARCM-M002204_seg001 | fru-Gal4 | M | 32482.08 | 1.00 |
| Gad1-F-200127 | 3973 | GadMARCM-F000596_seg003 | Gad1-Gal4 | F | 17090.43 | 0.54 |
| Gad1-F-100072 | 4001 | GadMARCM-F000618_seg001 | Gad1-Gal4 | F | 13951.95 | 0.53 |
| Gad1-F-900052 | 4034 | GadMARCM-F000639_seg001 | Gad1-Gal4 | F | 20465.14 | 0.57 |
| Gad1-F-100077 | 4061 | GadMARCM-F000658_seg001 | Gad1-Gal4 | F | 15723.05 | 0.52 |
| Gad1-F-000079 | 4079 | GadMARCM-F000669_seg001 | Gad1-Gal4 | F | 19144.16 | 0.66 |
| fru-F-400180 | 4693 | FruMARCM-F001928_seg001 | fru-Gal4 | F | 16530.99 | 0.58 |
| Cha-F-100221 | 5055 | ChaMARCM-F001132_seg001 | Cha-Gal4 | F | 17314.33 | 0.62 |
| Cha-F-000242 | 5106 | ChaMARCM-F001163_seg001 | Cha-Gal4 | F | 18527.50 | 0.54 |
| Cha-F-300186 | 5274 | ChaMARCM-F000669_seg001 | Cha-Gal4 | F | 17563.71 | 0.64 |
| Cha-F-300221 | 5476 | ChaMARCM-F000804_seg001 | Cha-Gal4 | F | 16365.18 | 0.60 |
| Cha-F-200186 | 5654 | ChaMARCM-F000926_seg001 | Cha-Gal4 | F | 15330.54 | 0.48 |
| VGlut-F-300393 | 6109 | DvGlutMARCM-F2181_seg1 | VGlut-Gal4 | F | 18385.31 | 0.63 |
| VGlut-F-200382 | 6115 | DvGlutMARCM-F2194_seg1 | VGlut-Gal4 | F | 22645.54 | 0.67 |
| VGlut-F-700430 | 7386 | DvGlutMARCM-F003842_seg002 | VGlut-Gal4 | F | 17982.22 | 0.65 |
| VGlut-F-700436 | 7392 | DvGlutMARCM-F003852_seg001 | VGlut-Gal4 | F | 16489.15 | 0.60 |
| Cha-F-100028 | 7523 | ChaMARCM-F000374_seg003 | Cha-Gal4 | F | 17713.52 | 0.61 |
| Cha-F-600081 | 7721 | ChaMARCM-F000533_seg001 | Cha-Gal4 | F | 19534.54 | 0.67 |
| Gad1-F-500038 | 10656 | GadMARCM-F000168_seg001 | Gad1-Gal4 | F | 19996.69 | 0.68 |
| VGlut-F-200389 | 11702 | DvGlutMARCM-F002275_seg001 | VGlut-Gal4 | F | 14648.74 | 0.57 |
| VGlut-F-500444 | 12190 | DvGlutMARCM-F002664_seg002 | VGlut-Gal4 | F | 15921.38 | 0.57 |
| VGlut-F-900028 | 12874 | DvGlutMARCM-F1915_seg1 | VGlut-Gal4 | F | 16629.62 | 0.55 |
| VGlut-F-500323 | 13029 | DvGlutMARCM-F2054_seg1 | VGlut-Gal4 | F | 16556.37 | 0.53 |
| VGlut-F-500253 | 15406 | DvGlutMARCM-F1764_seg1 | VGlut-Gal4 | F | 17067.52 | 0.60 |
| VGlut-F-200021 | 15627 | DvGlutMARCM-F043-X3_seg2 | VGlut-Gal4 | F | 22507.05 | 0.67 |