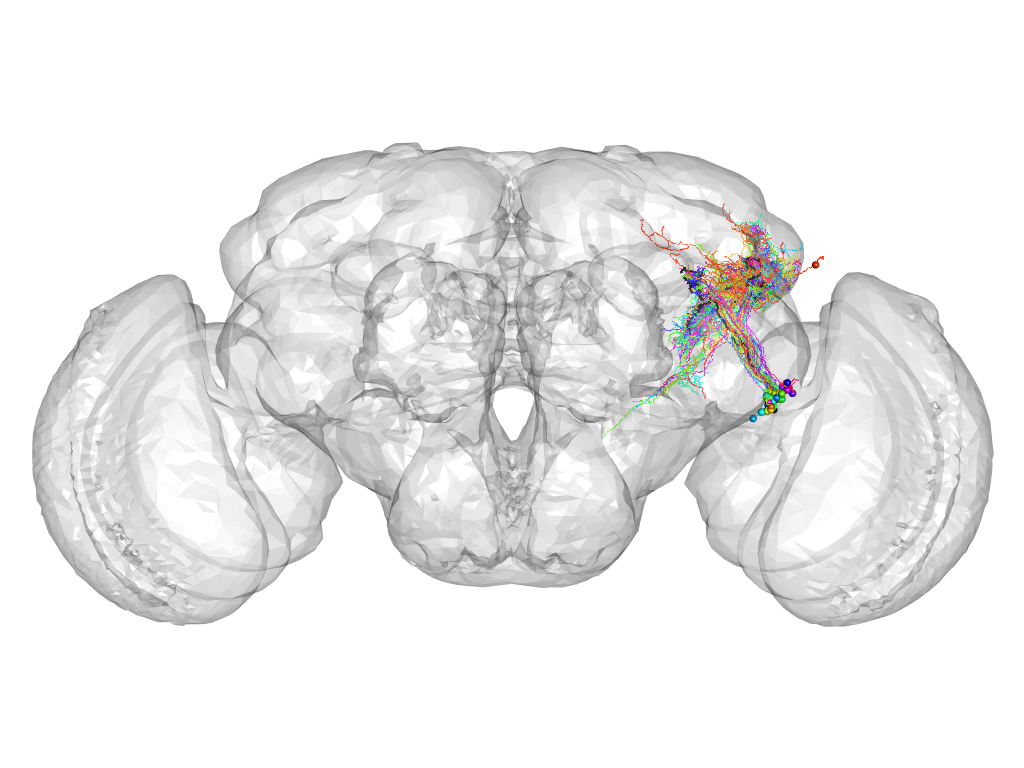
Cluster 998
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-100009), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 674 | Gad1-F-600005 | X | 0.68 |
| 662 | Tdc2-F-400026 | XIII | 0.58 |
| 621 | Gad1-F-500089 | XVII | 0.57 |
| 2 | fru-M-200253 | XVII | 0.56 |
| 339 | Cha-F-100223 | XVII | 0.54 |
| 872 | TH-F-100101 | IX | 0.54 |
| 136 | TH-M-000013 | XVII | 0.54 |
Cluster composition
This cluster has 37 neurons. The exemplar of this cluster (VGlut-F-100009) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-100009 | 15761 | DvGlutMARCM-F168_seg1 | VGlut-Gal4 | F | 51912.11 | 1.00 |
| fru-M-600094 | 48 | FruMARCM-M002241_seg001 | fru-Gal4 | M | 33515.44 | 0.58 |
| fru-M-300124 | 971 | FruMARCM-M001146_seg001 | fru-Gal4 | M | 30902.63 | 0.52 |
| fru-M-100125 | 1159 | FruMARCM-M001347_seg003 | fru-Gal4 | M | 33712.16 | 0.63 |
| fru-F-600103 | 4593 | FruMARCM-F001710_seg001 | fru-Gal4 | F | 35515.96 | 0.60 |
| VGlut-F-700565 | 5750 | DvGlutMARCM-F004455_seg002 | VGlut-Gal4 | F | 23702.19 | 0.48 |
| VGlut-F-500847 | 5759 | DvGlutMARCM-F004463_seg001 | VGlut-Gal4 | F | 35013.83 | 0.66 |
| VGlut-F-900129 | 6640 | DvGlutMARCM-F004388_seg001 | VGlut-Gal4 | F | 28749.48 | 0.61 |
| VGlut-F-500744 | 6742 | DvGlutMARCM-F003947_seg001 | VGlut-Gal4 | F | 35633.78 | 0.69 |
| VGlut-F-600713 | 6785 | DvGlutMARCM-F003980_seg001 | VGlut-Gal4 | F | 29015.94 | 0.62 |
| VGlut-F-600739 | 6880 | DvGlutMARCM-F004038_seg005 | VGlut-Gal4 | F | 33862.95 | 0.62 |
| VGlut-F-600773 | 7064 | DvGlutMARCM-F004193_seg001 | VGlut-Gal4 | F | 33315.35 | 0.55 |
| VGlut-F-700266 | 8500 | DvGlutMARCM-F003083_seg001 | VGlut-Gal4 | F | 36013.79 | 0.71 |
| VGlut-F-700284 | 8521 | DvGlutMARCM-F003100_seg001 | VGlut-Gal4 | F | 37174.25 | 0.69 |
| VGlut-F-600516 | 8548 | DvGlutMARCM-F003119_seg001 | VGlut-Gal4 | F | 33904.82 | 0.67 |
| VGlut-F-600598 | 9045 | DvGlutMARCM-F003368_seg001 | VGlut-Gal4 | F | 36810.07 | 0.71 |
| VGlut-F-500603 | 9384 | DvGlutMARCM-F003214_seg001 | VGlut-Gal4 | F | 31936.67 | 0.62 |
| Gad1-F-300038 | 10566 | GadMARCM-F000089_seg001 | Gad1-Gal4 | F | 34963.07 | 0.59 |
| VGlut-F-400241 | 10756 | DvGlutMARCM-F843_seg1 | VGlut-Gal4 | F | 36508.69 | 0.72 |
| VGlut-F-500507 | 11160 | DvGlutMARCM-F003011_seg002 | VGlut-Gal4 | F | 34185.95 | 0.68 |
| Cha-F-500001 | 11578 | ChaMARCM-F000014_seg002 | Cha-Gal4 | F | 32797.47 | 0.55 |
| VGlut-F-300442 | 11720 | DvGlutMARCM-F002293_seg001 | VGlut-Gal4 | F | 29572.88 | 0.63 |
| VGlut-F-600228 | 11830 | DvGlutMARCM-F002389_seg001 | VGlut-Gal4 | F | 35316.88 | 0.68 |
| VGlut-F-400535 | 11913 | DvGlutMARCM-F002448_seg002 | VGlut-Gal4 | F | 35255.71 | 0.71 |
| VGlut-F-500378 | 11922 | DvGlutMARCM-F002457_seg001 | VGlut-Gal4 | F | 33907.02 | 0.67 |
| VGlut-F-500269 | 12787 | DvGlutMARCM-F1835_seg1 | VGlut-Gal4 | F | 33894.88 | 0.65 |
| VGlut-F-600109 | 12932 | DvGlutMARCM-F1970_seg3 | VGlut-Gal4 | F | 39852.75 | 0.75 |
| VGlut-F-400166 | 14419 | DvGlutMARCM-F713_seg2 | VGlut-Gal4 | F | 30350.62 | 0.66 |
| VGlut-F-500113 | 14530 | DvGlutMARCM-F800-x2_seg1 | VGlut-Gal4 | F | 34379.90 | 0.68 |
| VGlut-F-600064 | 15118 | DvGlutMARCM-F1451_seg2 | VGlut-Gal4 | F | 32144.04 | 0.57 |
| VGlut-F-600069 | 15148 | DvGlutMARCM-F1483_seg1 | VGlut-Gal4 | F | 32376.26 | 0.61 |
| VGlut-F-600072 | 15151 | DvGlutMARCM-F1484_seg2 | VGlut-Gal4 | F | 38163.03 | 0.72 |
| VGlut-F-000366 | 15187 | DvGlutMARCM-F1517_seg1 | VGlut-Gal4 | F | 32455.36 | 0.64 |
| VGlut-F-600078 | 15267 | DvGlutMARCM-F1589_seg1 | VGlut-Gal4 | F | 37497.69 | 0.71 |
| VGlut-F-600081 | 15296 | DvGlutMARCM-F1629_seg1 | VGlut-Gal4 | F | 27557.28 | 0.56 |
| VGlut-F-500255 | 15408 | DvGlutMARCM-F1766_seg1 | VGlut-Gal4 | F | 35463.73 | 0.67 |
| VGlut-F-700009 | 15737 | DvGlutMARCM-F148_seg1 | VGlut-Gal4 | F | 36974.50 | 0.72 |