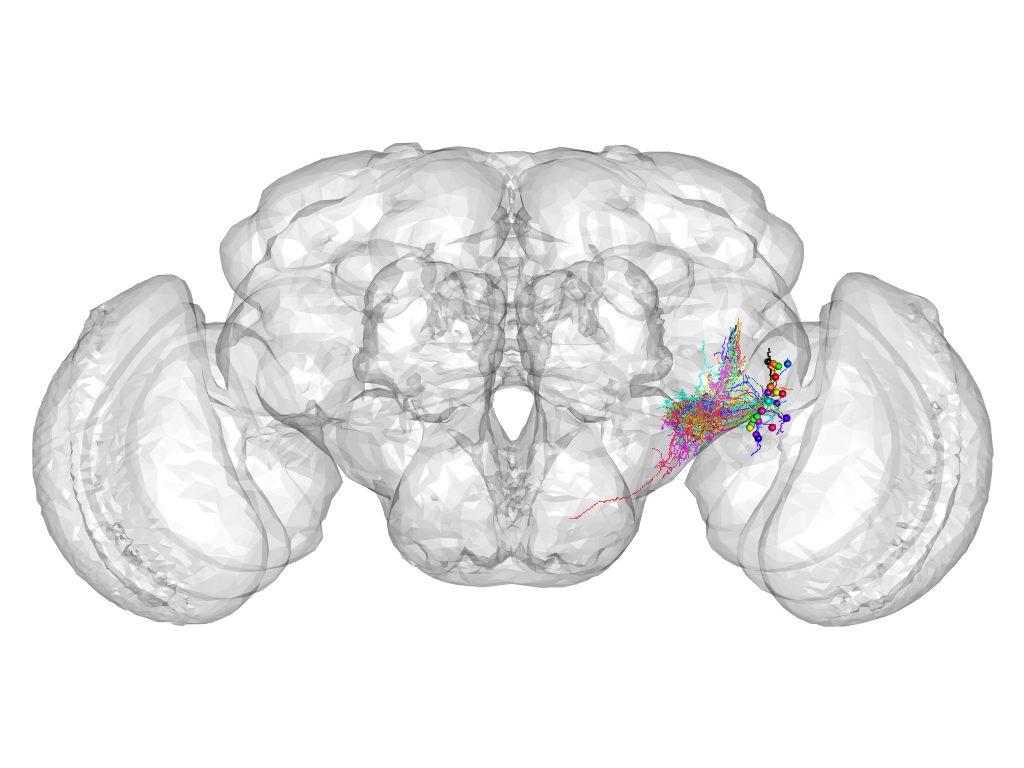
Cluster 474
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-700092), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 829 | VGlut-F-000436 | X | 0.71 |
| 200 | TH-M-000060 | X | 0.71 |
| 251 | Gad1-F-800086 | X | 0.69 |
| 903 | VGlut-F-000273 | X | 0.66 |
| 146 | Trh-M-200088 | VIII | 0.60 |
| 532 | Cha-F-300093 | X | 0.60 |
| 865 | TH-F-000025 | X | 0.55 |
| 652 | Tdc2-F-200010 | VIII | 0.53 |
| 148 | Trh-M-400144 | X | 0.52 |
| 876 | Trh-F-100023 | X | 0.52 |
Cluster composition
This cluster has 26 neurons. The exemplar of this cluster (fru-F-700092) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-700092 | 7327 | FruMARCM-F000965_seg001 | fru-Gal4 | F | 7642.17 | 1.00 |
| fru-M-800086 | 920 | FruMARCM-M002181_seg001 | fru-Gal4 | M | 5669.03 | 0.63 |
| fru-M-800087 | 921 | FruMARCM-M002181_seg002 | fru-Gal4 | M | 5159.34 | 0.67 |
| fru-M-500060 | 1366 | FruMARCM-M000876_seg001 | fru-Gal4 | M | 5579.74 | 0.75 |
| fru-M-600034 | 1408 | FruMARCM-M000906_seg002 | fru-Gal4 | M | 5827.00 | 0.77 |
| fru-M-400043 | 1568 | FruMARCM-M000640_seg001 | fru-Gal4 | M | 5209.45 | 0.70 |
| fru-M-400044 | 1569 | FruMARCM-M000641_seg001 | fru-Gal4 | M | 5348.38 | 0.70 |
| Trh-M-600086 | 2030 | TPHMARCM-M001598_seg001 | Trh-Gal4 | M | 4986.07 | 0.68 |
| Trh-M-500179 | 2084 | TPHMARCM-M001585_seg001 | Trh-Gal4 | M | 5155.54 | 0.69 |
| fru-M-600003 | 2411 | FruMARCM-M000132_seg001 | fru-Gal4 | M | 5381.22 | 0.73 |
| Trh-M-400069 | 2837 | TPHMARCM-1063M_seg1 | Trh-Gal4 | M | 4986.84 | 0.67 |
| Cha-F-200273 | 4138 | ChaMARCM-F001255_seg001 | Cha-Gal4 | F | 5407.45 | 0.71 |
| Cha-F-100266 | 4211 | ChaMARCM-F001301_seg001 | Cha-Gal4 | F | 5172.90 | 0.67 |
| fru-F-500187 | 6325 | FruMARCM-F001446_seg003 | fru-Gal4 | F | 4886.25 | 0.62 |
| fru-F-500189 | 6374 | FruMARCM-F001511_seg002 | fru-Gal4 | F | 5671.11 | 0.76 |
| Cha-F-100037 | 7242 | ChaMARCM-F000415_seg001 | Cha-Gal4 | F | 5109.79 | 0.65 |
| fru-F-800035 | 7300 | FruMARCM-F000929_seg001 | fru-Gal4 | F | 5264.58 | 0.71 |
| fru-F-500134 | 7818 | FruMARCM-F000712_seg001 | fru-Gal4 | F | 5655.18 | 0.73 |
| fru-F-400044 | 9751 | FruMARCM-F000265_seg001 | fru-Gal4 | F | 5039.61 | 0.65 |
| fru-F-400065 | 9837 | FruMARCM-F000405_seg001 | fru-Gal4 | F | 5473.63 | 0.69 |
| fru-F-500037 | 9947 | FruMARCM-F000541_seg001 | fru-Gal4 | F | 5510.75 | 0.75 |
| Cha-F-300001 | 11571 | ChaMARCM-F000009_seg002 | Cha-Gal4 | F | 4541.83 | 0.57 |
| Trh-F-500168 | 12512 | TPHMARCM-1294F_seg1 | Trh-Gal4 | F | 4914.65 | 0.65 |
| Trh-F-400010 | 13846 | TPHMARCM-115F_seg1 | Trh-Gal4 | F | 4955.15 | 0.66 |
| Trh-F-400015 | 13871 | TPHMARCM-140F_seg2 | Trh-Gal4 | F | 3932.99 | 0.50 |
| Trh-F-700041 | 14295 | TPHMARCM-836F_seg1 | Trh-Gal4 | F | 5037.64 | 0.68 |