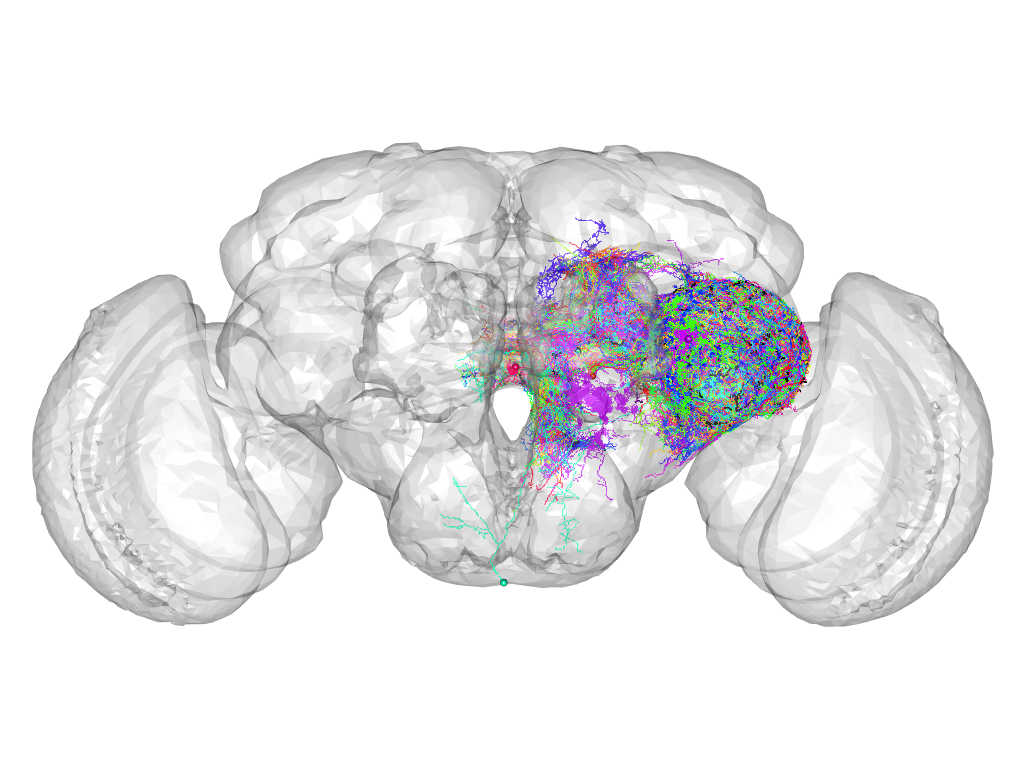
Cluster 876
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-100023), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 200 | TH-M-000060 | X | 0.71 |
| 939 | VGlut-F-400341 | X | 0.61 |
| 865 | TH-F-000025 | X | 0.60 |
| 1038 | VGlut-F-000111 | X | 0.46 |
| 1031 | VGlut-F-400089 | X | 0.46 |
Cluster composition
This cluster has 45 neurons. The exemplar of this cluster (Trh-F-100023) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-100023 | 13817 | TPHMARCM-036F_seg1 | Trh-Gal4 | F | 348966.00 | 1.00 |
| fru-M-200329 | 348 | FruMARCM-M002581_seg001 | fru-Gal4 | M | 254965.21 | 0.71 |
| fru-M-800079 | 713 | FruMARCM-M001917_seg001 | fru-Gal4 | M | 236100.74 | 0.70 |
| fru-M-800029 | 1673 | FruMARCM-M000830_seg001 | fru-Gal4 | M | 244404.11 | 0.71 |
| Trh-M-000140 | 1732 | TPHMARCM-M001809_seg001 | Trh-Gal4 | M | 248614.88 | 0.72 |
| Trh-M-000144 | 1736 | TPHMARCM-M001813_seg001 | Trh-Gal4 | M | 245721.22 | 0.71 |
| Trh-M-000148 | 1740 | TPHMARCM-M001817_seg001 | Trh-Gal4 | M | 249383.89 | 0.73 |
| Trh-M-000152 | 1743 | TPHMARCM-M001820_seg001 | Trh-Gal4 | M | 253274.69 | 0.72 |
| Trh-M-000077 | 2247 | TPHMARCM-M001734_seg001 | Trh-Gal4 | M | 252353.24 | 0.73 |
| Trh-M-000105 | 2291 | TPHMARCM-M001773_seg002 | Trh-Gal4 | M | 255885.35 | 0.74 |
| Trh-M-000119 | 2305 | TPHMARCM-M001787_seg001 | Trh-Gal4 | M | 246975.95 | 0.71 |
| Trh-M-000123 | 2309 | TPHMARCM-M001791_seg001 | Trh-Gal4 | M | 246398.24 | 0.71 |
| Trh-M-000023 | 2649 | TPHMARCM-M001418_seg001 | Trh-Gal4 | M | 246928.48 | 0.71 |
| TH-M-100009 | 3101 | THMARCM-079M_seg1 | TH-Gal4 | M | 244592.31 | 0.70 |
| TH-M-100023 | 3154 | THMARCM-204M_seg1 | TH-Gal4 | M | 250093.47 | 0.72 |
| TH-M-100027 | 3159 | THMARCM-216M_seg1 | TH-Gal4 | M | 241576.98 | 0.72 |
| TH-M-100037 | 3179 | THMARCM-271M_seg1 | TH-Gal4 | M | 242069.15 | 0.69 |
| TH-M-500005 | 3343 | THMARCM-633M_seg | TH-Gal4 | M | 244071.51 | 0.70 |
| Trh-M-000001 | 3386 | TPHMARCM-063M_seg1 | Trh-Gal4 | M | 256073.86 | 0.74 |
| VGlut-F-400756 | 8655 | DvGlutMARCM-F003704_seg001 | VGlut-Gal4 | F | 245545.84 | 0.71 |
| fru-F-300032 | 9843 | FruMARCM-F000428_seg001 | fru-Gal4 | F | 255157.27 | 0.74 |
| Gad1-F-000013 | 10667 | GadMARCM-F000175_seg001 | Gad1-Gal4 | F | 198595.07 | 0.56 |
| VGlut-F-900062 | 11072 | DvGlutMARCM-F002951_seg001 | VGlut-Gal4 | F | 221595.12 | 0.70 |
| Trh-F-100068 | 12693 | TPHMARCM-1108F_seg1 | Trh-Gal4 | F | 258583.85 | 0.74 |
| Trh-F-100069 | 12694 | TPHMARCM-1109F_seg1 | Trh-Gal4 | F | 247697.89 | 0.73 |
| Trh-F-000031 | 13300 | TPHMARCM-782F_seg1 | Trh-Gal4 | F | 247560.65 | 0.71 |
| TH-F-000009 | 13344 | THMARCM-098F_seg1 | TH-Gal4 | F | 239749.89 | 0.69 |
| TH-F-300012 | 13389 | THMARCM-157F_seg1 | TH-Gal4 | F | 251530.91 | 0.72 |
| TH-F-100023 | 13426 | THMARCM-237F_seg1 | TH-Gal4 | F | 253573.71 | 0.73 |
| TH-F-100024 | 13427 | THMARCM-238F_seg1 | TH-Gal4 | F | 243027.12 | 0.72 |
| TH-F-100026 | 13429 | THMARCM-241F_seg1 | TH-Gal4 | F | 225066.20 | 0.67 |
| TH-F-200009 | 13433 | THMARCM-249F_seg1 | TH-Gal4 | F | 245887.94 | 0.71 |
| TH-F-200020 | 13447 | THMARCM-285F_seg1 | TH-Gal4 | F | 265518.89 | 0.72 |
| TH-F-200045 | 13489 | THMARCM-350F_seg1 | TH-Gal4 | F | 245532.81 | 0.69 |
| TH-F-000040 | 13645 | THMARCM-649F_seg | TH-Gal4 | F | 237364.01 | 0.70 |
| TH-F-000073 | 13721 | THMARCM-751F_seg1 | TH-Gal4 | F | 254236.07 | 0.72 |
| TH-F-000079 | 13734 | THMARCM-762F_seg2 | TH-Gal4 | F | 252188.44 | 0.65 |
| TH-F-100109 | 13755 | THMARCM-792F_seg1 | TH-Gal4 | F | 239378.88 | 0.71 |
| TH-F-000100 | 13766 | THMARCM-818F_seg1 | TH-Gal4 | F | 245300.06 | 0.71 |
| Trh-F-000001 | 13798 | TPHMARCM-016F_seg1 | Trh-Gal4 | F | 236839.72 | 0.71 |
| Trh-F-300072 | 14131 | TPHMARCM-521F_seg1 | Trh-Gal4 | F | 249531.55 | 0.72 |
| VGlut-F-000232 | 14706 | DvGlutMARCM-F963_seg1 | VGlut-Gal4 | F | 251808.01 | 0.72 |
| VGlut-F-000347 | 15037 | DvGlutMARCM-F1375_seg1 | VGlut-Gal4 | F | 250984.40 | 0.72 |
| VGlut-F-300246 | 15075 | DvGlutMARCM-F1413_seg1 | VGlut-Gal4 | F | 242504.47 | 0.70 |
| VGlut-F-200042 | 15660 | DvGlutMARCM-F072_seg1 | VGlut-Gal4 | F | 245826.66 | 0.71 |