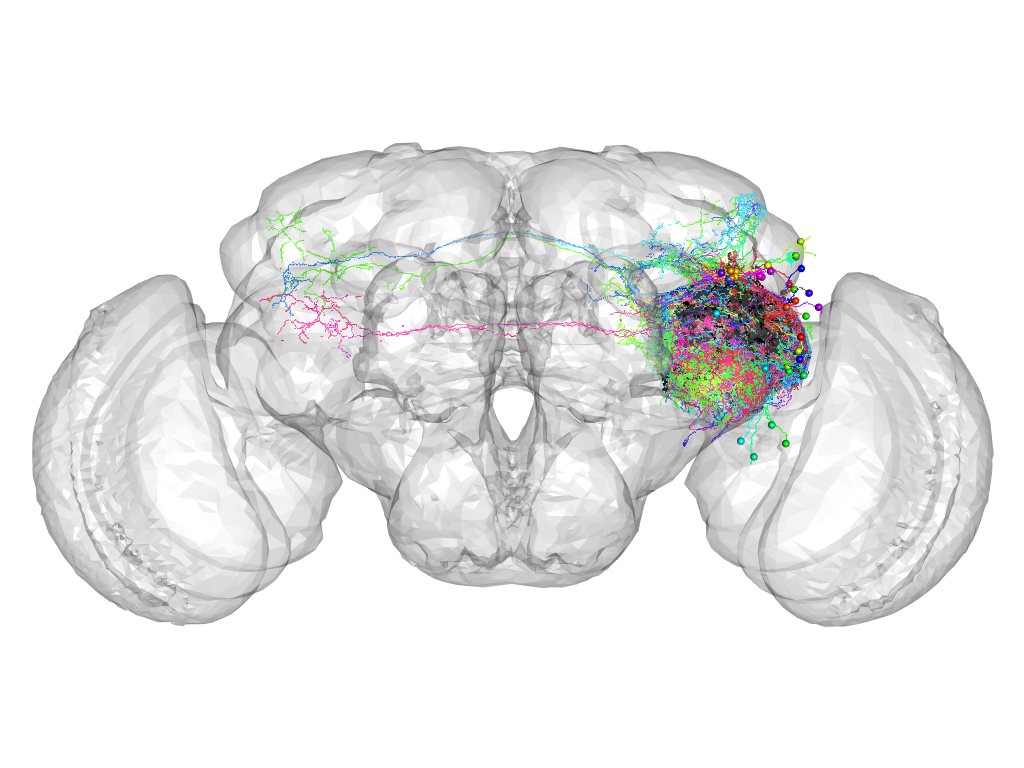
Cluster 939
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-400341), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 876 | Trh-F-100023 | X | 0.74 |
| 200 | TH-M-000060 | X | 0.70 |
| 1038 | VGlut-F-000111 | X | 0.66 |
| 865 | TH-F-000025 | X | 0.61 |
| 321 | Cha-F-600151 | X | 0.52 |
| 1031 | VGlut-F-400089 | X | 0.51 |
Cluster composition
This cluster has 42 neurons. The exemplar of this cluster (VGlut-F-400341) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-400341 | 14878 | DvGlutMARCM-F1222_seg1 | VGlut-Gal4 | F | 487777.93 | 1.00 |
| fru-M-200328 | 347 | FruMARCM-M002580_seg001 | fru-Gal4 | M | 149904.85 | 0.50 |
| fru-M-800074 | 632 | FruMARCM-M001843_seg001 | fru-Gal4 | M | 66501.75 | 0.47 |
| fru-M-800013 | 2483 | FruMARCM-M000237_seg002 | fru-Gal4 | M | 235706.83 | 0.60 |
| Trh-M-500138 | 2735 | TPHMARCM-M001502_seg002 | Trh-Gal4 | M | 183286.59 | 0.55 |
| Trh-M-400104 | 2882 | TPHMARCM-1124M_seg1 | Trh-Gal4 | M | 257826.06 | 0.66 |
| Trh-M-400045 | 2998 | TPHMARCM-941M_seg1 | Trh-Gal4 | M | 269700.72 | 0.67 |
| Trh-M-500000 | 3427 | TPHMARCM-134M_seg1 | Trh-Gal4 | M | 273338.05 | 0.68 |
| Trh-M-700010 | 3682 | TPHMARCM-794M_seg3 | Trh-Gal4 | M | 268088.75 | 0.68 |
| fru-F-500275 | 3772 | FruMARCM-F002397_seg001 | fru-Gal4 | F | 185623.89 | 0.52 |
| Cha-F-400237 | 3849 | ChaMARCM-F001521_seg001 | Cha-Gal4 | F | 341069.41 | 0.74 |
| Gad1-F-900060 | 4042 | GadMARCM-F000644_seg001 | Gad1-Gal4 | F | 273383.99 | 0.65 |
| Cha-F-200299 | 4219 | ChaMARCM-F001308_seg001 | Cha-Gal4 | F | 314853.40 | 0.67 |
| Cha-F-100332 | 4383 | ChaMARCM-F001436_seg002 | Cha-Gal4 | F | 351855.65 | 0.62 |
| Cha-F-000107 | 5251 | ChaMARCM-F000652_seg001 | Cha-Gal4 | F | 158764.76 | 0.45 |
| Cha-F-800031 | 5411 | ChaMARCM-F000764_seg002 | Cha-Gal4 | F | 199177.04 | 0.50 |
| Cha-F-600108 | 5424 | ChaMARCM-F000774_seg002 | Cha-Gal4 | F | 236376.74 | 0.61 |
| Cha-F-000144 | 5482 | ChaMARCM-F000808_seg002 | Cha-Gal4 | F | 290588.63 | 0.68 |
| VGlut-F-200592 | 5719 | DvGlutMARCM-F004426_seg002 | VGlut-Gal4 | F | 213981.33 | 0.58 |
| Cha-F-700002 | 6144 | ChaMARCM-F000028_seg001 | Cha-Gal4 | F | 216681.93 | 0.58 |
| VGlut-F-300594 | 7015 | DvGlutMARCM-F004154_seg001 | VGlut-Gal4 | F | 267190.98 | 0.62 |
| Cha-F-200075 | 7595 | ChaMARCM-F000447_seg001 | Cha-Gal4 | F | 186328.60 | 0.54 |
| Cha-F-200090 | 7646 | ChaMARCM-F000478_seg001 | Cha-Gal4 | F | 198497.69 | 0.44 |
| Cha-F-000087 | 7695 | ChaMARCM-F000514_seg001 | Cha-Gal4 | F | 257936.86 | 0.67 |
| fru-F-900007 | 9797 | FruMARCM-F000337_seg002 | fru-Gal4 | F | 90585.59 | 0.40 |
| Cha-F-300019 | 10103 | ChaMARCM-F000057_seg002 | Cha-Gal4 | F | 263709.43 | 0.46 |
| Cha-F-500015 | 10114 | ChaMARCM-F000066_seg002 | Cha-Gal4 | F | 203938.95 | 0.41 |
| Trh-F-000057 | 10457 | TPHMARCM-F001401_seg002 | Trh-Gal4 | F | 290468.26 | 0.69 |
| Gad1-F-300052 | 10701 | GadMARCM-F000199_seg002 | Gad1-Gal4 | F | 138106.43 | 0.51 |
| Gad1-F-500006 | 10734 | GadMARCM-F000059_seg001 | Gad1-Gal4 | F | 249956.51 | 0.61 |
| Cha-F-400003 | 11588 | ChaMARCM-F000020_seg003 | Cha-Gal4 | F | 342735.59 | 0.74 |
| Trh-F-500167 | 12511 | TPHMARCM-1293F_seg1 | Trh-Gal4 | F | 307720.84 | 0.68 |
| Trh-F-700047 | 12717 | TPHMARCM-879F_seg2 | Trh-Gal4 | F | 310037.24 | 0.72 |
| VGlut-F-100230 | 13125 | DvGlutMARCM-F2151_seg1 | VGlut-Gal4 | F | 272589.67 | 0.66 |
| Trh-F-400070 | 13226 | TPHMARCM-713F_seg1 | Trh-Gal4 | F | 214283.59 | 0.57 |
| Trh-F-500016 | 13863 | TPHMARCM-131F_seg2 | Trh-Gal4 | F | 292491.65 | 0.69 |
| Trh-F-400046 | 14065 | TPHMARCM-403F_seg1 | Trh-Gal4 | F | 291552.71 | 0.70 |
| VGlut-F-000266 | 14345 | DvGlutMARCM-F1029_seg1 | VGlut-Gal4 | F | 290643.85 | 0.65 |
| VGlut-F-300201 | 14745 | DvGlutMARCM-F1096_seg1 | VGlut-Gal4 | F | 263419.84 | 0.63 |
| VGlut-F-000323 | 14879 | DvGlutMARCM-F1223_seg1 | VGlut-Gal4 | F | 327111.03 | 0.63 |
| VGlut-F-200265 | 14984 | DvGlutMARCM-F1323_seg1 | VGlut-Gal4 | F | 268331.55 | 0.66 |
| VGlut-F-000016 | 15742 | DvGlutMARCM-F153-X2_seg1 | VGlut-Gal4 | F | 379547.74 | 0.77 |