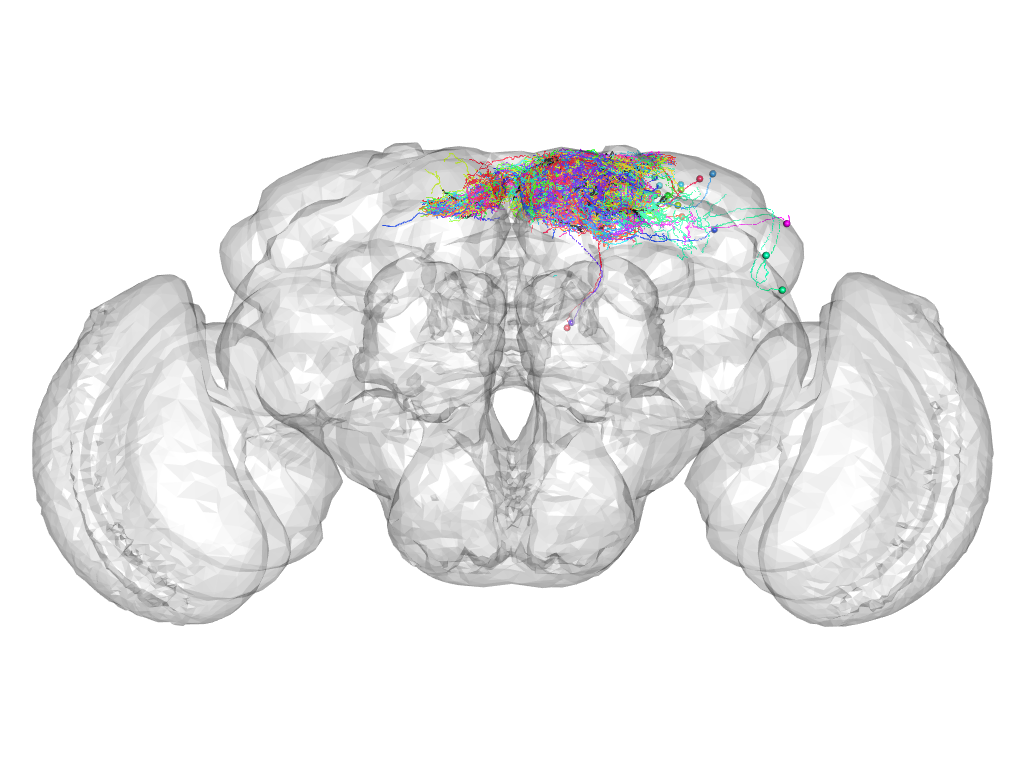
Cluster 40
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-000123), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 881 | Trh-F-200023 | XIX | 0.67 |
| 95 | fru-M-000064 | XIX | 0.59 |
| 206 | Trh-M-000018 | XIX | 0.56 |
| 15 | fru-M-300268 | XIX | 0.56 |
| 1003 | VGlut-F-000024 | XIX | 0.55 |
| 199 | TH-M-000044 | XIX | 0.55 |
| 117 | fru-M-500059 | XVIII | 0.53 |
| 850 | TH-F-100027 | XIX | 0.52 |
| 39 | fru-M-000219 | XIX | 0.50 |
Cluster composition
This cluster has 50 neurons. The exemplar of this cluster (fru-M-000123) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-000123 | 415 | FruMARCM-M001907_seg001 | fru-Gal4 | M | 39406.73 | 1.00 |
| fru-M-200255 | 38 | FruMARCM-M002234_seg001 | fru-Gal4 | M | 29543.87 | 0.73 |
| fru-M-900062 | 171 | FruMARCM-M002389_seg001 | fru-Gal4 | M | 28950.31 | 0.72 |
| fru-M-700177 | 260 | FruMARCM-M002486_seg001 | fru-Gal4 | M | 30011.70 | 0.74 |
| fru-M-000207 | 336 | FruMARCM-M002556_seg003 | fru-Gal4 | M | 29350.16 | 0.75 |
| fru-M-500239 | 339 | FruMARCM-M002573_seg001 | fru-Gal4 | M | 29850.63 | 0.74 |
| fru-M-100225 | 354 | FruMARCM-M002588_seg001 | fru-Gal4 | M | 30063.97 | 0.76 |
| fru-M-100147 | 457 | FruMARCM-M001566_seg001 | fru-Gal4 | M | 28584.69 | 0.74 |
| fru-M-500163 | 570 | FruMARCM-M001755_seg002 | fru-Gal4 | M | 28537.95 | 0.75 |
| fru-M-600082 | 630 | FruMARCM-M001841_seg001 | fru-Gal4 | M | 27484.88 | 0.71 |
| fru-M-300208 | 644 | FruMARCM-M001856_seg001 | fru-Gal4 | M | 29685.06 | 0.75 |
| fru-M-300214 | 661 | FruMARCM-M001871_seg002 | fru-Gal4 | M | 29441.01 | 0.73 |
| fru-M-600087 | 726 | FruMARCM-M001936_seg001 | fru-Gal4 | M | 29214.32 | 0.74 |
| fru-M-100188 | 840 | FruMARCM-M002054_seg001 | fru-Gal4 | M | 30162.92 | 0.77 |
| fru-M-500222 | 895 | FruMARCM-M002163_seg001 | fru-Gal4 | M | 29482.96 | 0.75 |
| fru-M-500105 | 988 | FruMARCM-M001170_seg001 | fru-Gal4 | M | 27997.94 | 0.72 |
| fru-M-200151 | 1015 | FruMARCM-M001198_seg001 | fru-Gal4 | M | 27808.80 | 0.70 |
| fru-M-100098 | 1036 | FruMARCM-M001220_seg001 | fru-Gal4 | M | 28025.64 | 0.73 |
| fru-M-400129 | 1117 | FruMARCM-M001316_seg001 | fru-Gal4 | M | 27725.89 | 0.72 |
| fru-M-200201 | 1293 | FruMARCM-M001486_seg001 | fru-Gal4 | M | 28123.05 | 0.73 |
| fru-M-300095 | 1358 | FruMARCM-M000869_seg001 | fru-Gal4 | M | 29466.54 | 0.74 |
| fru-M-400081 | 1449 | FruMARCM-M001017_seg002 | fru-Gal4 | M | 27600.99 | 0.71 |
| npf-M-200001 | 1751 | npfMARCM-M000051_seg001 | npf-GAL4 | M | 23623.96 | 0.52 |
| npf-M-200002 | 1752 | npfMARCM-M000052_seg001 | npf-GAL4 | M | 22834.46 | 0.52 |
| fru-M-100028 | 1775 | FruMARCM-M000292_seg001 | fru-Gal4 | M | 27655.83 | 0.72 |
| fru-M-400035 | 1884 | FruMARCM-M000422_seg001 | fru-Gal4 | M | 27419.80 | 0.71 |
| fru-M-200021 | 2340 | FruMARCM-M000030_seg006 | fru-Gal4 | M | 27208.43 | 0.70 |
| fru-M-800000 | 2363 | FruMARCM-M000097_seg001 | fru-Gal4 | M | 26430.81 | 0.71 |
| fru-F-200175 | 3702 | FruMARCM-F002566_seg001 | fru-Gal4 | F | 29169.35 | 0.74 |
| fru-F-100087 | 3795 | FruMARCM-F002472_seg001 | fru-Gal4 | F | 29979.74 | 0.74 |
| fru-F-000144 | 3826 | FruMARCM-F002637_seg001 | fru-Gal4 | F | 30226.58 | 0.75 |
| fru-F-000145 | 3828 | FruMARCM-F002652_seg001 | fru-Gal4 | F | 29628.18 | 0.75 |
| Cha-F-700177 | 4446 | ChaMARCM-F001489_seg001 | Cha-Gal4 | F | 21490.96 | 0.58 |
| fru-F-800049 | 4579 | FruMARCM-F001695_seg001 | fru-Gal4 | F | 28216.50 | 0.71 |
| fru-F-800050 | 4590 | FruMARCM-F001706_seg001 | fru-Gal4 | F | 28084.46 | 0.71 |
| fru-F-100074 | 4815 | FruMARCM-F002182_seg001 | fru-Gal4 | F | 28924.29 | 0.74 |
| Cha-F-400152 | 5304 | ChaMARCM-F000687_seg001 | Cha-Gal4 | F | 20509.40 | 0.57 |
| Cha-F-300242 | 5540 | ChaMARCM-F000844_seg002 | Cha-Gal4 | F | 26179.35 | 0.66 |
| fru-F-500159 | 6178 | FruMARCM-F001112_seg001 | fru-Gal4 | F | 26932.05 | 0.68 |
| fru-F-400139 | 6230 | FruMARCM-F001181_seg001 | fru-Gal4 | F | 26184.70 | 0.68 |
| fru-F-600084 | 6385 | FruMARCM-F001522_seg001 | fru-Gal4 | F | 28708.37 | 0.73 |
| fru-F-400118 | 7338 | FruMARCM-F000975_seg001 | fru-Gal4 | F | 27767.46 | 0.71 |
| Cha-F-700122 | 7642 | ChaMARCM-F000475_seg001 | Cha-Gal4 | F | 22677.07 | 0.57 |
| fru-F-800018 | 7832 | FruMARCM-F000733_seg001 | fru-Gal4 | F | 26212.89 | 0.70 |
| fru-F-200058 | 7841 | FruMARCM-F000750_seg001 | fru-Gal4 | F | 28259.58 | 0.73 |
| fru-F-300047 | 7865 | FruMARCM-F000779_seg001 | fru-Gal4 | F | 28424.75 | 0.73 |
| fru-F-000034 | 7895 | FruMARCM-F000825_seg002 | fru-Gal4 | F | 27289.87 | 0.71 |
| fru-F-700044 | 9925 | FruMARCM-F000505_seg001 | fru-Gal4 | F | 27159.04 | 0.72 |
| VGlut-F-300022 | 10752 | DvGlutMARCM-F226_seg1 | VGlut-Gal4 | F | 27642.87 | 0.72 |
| Gad1-F-400053 | 11399 | GadMARCM-F000349_seg002 | Gad1-Gal4 | F | 19341.89 | 0.49 |