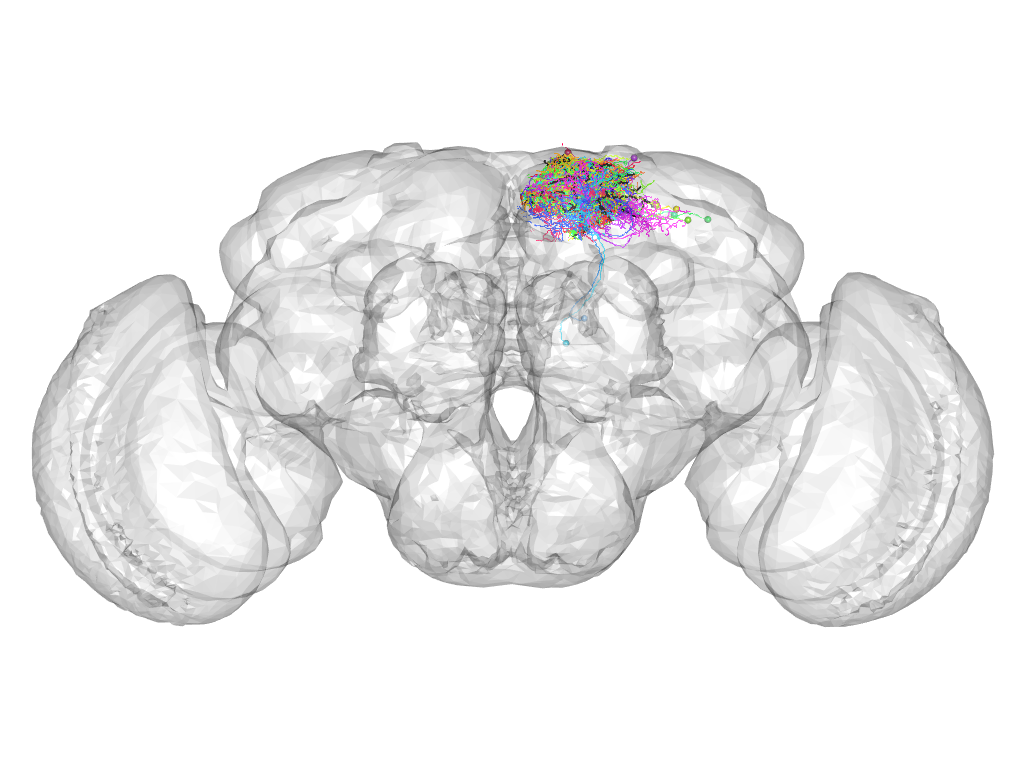
Cluster 15
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-300268), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 1003 | VGlut-F-000024 | XIX | 0.69 |
| 40 | fru-M-000123 | XIX | 0.68 |
| 881 | Trh-F-200023 | XIX | 0.62 |
| 193 | TH-M-100039 | XIX | 0.61 |
| 850 | TH-F-100027 | XIX | 0.59 |
| 206 | Trh-M-000018 | XIX | 0.58 |
| 886 | Trh-F-300042 | XIX | 0.58 |
| 95 | fru-M-000064 | XIX | 0.57 |
| 199 | TH-M-000044 | XIX | 0.57 |
| 39 | fru-M-000219 | XIX | 0.56 |
| 992 | VGlut-F-200024 | XIX | 0.51 |
Cluster composition
This cluster has 20 neurons. The exemplar of this cluster (fru-M-300268) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-300268 | 190 | FruMARCM-M002410_seg001 | fru-Gal4 | M | 27664.44 | 1.00 |
| fru-M-800098 | 209 | FruMARCM-M002424_seg001 | fru-Gal4 | M | 20355.06 | 0.75 |
| fru-M-000196 | 307 | FruMARCM-M002534_seg001 | fru-Gal4 | M | 21144.96 | 0.77 |
| fru-M-200313 | 316 | FruMARCM-M002539_seg002 | fru-Gal4 | M | 20502.71 | 0.75 |
| fru-M-000200 | 318 | FruMARCM-M002540_seg001 | fru-Gal4 | M | 21235.97 | 0.77 |
| fru-M-200315 | 319 | FruMARCM-M002541_seg001 | fru-Gal4 | M | 21002.91 | 0.76 |
| fru-M-000125 | 730 | FruMARCM-M001941_seg002 | fru-Gal4 | M | 21046.34 | 0.75 |
| fru-M-700134 | 824 | FruMARCM-M002029_seg001 | fru-Gal4 | M | 19715.73 | 0.73 |
| fru-M-500216 | 861 | FruMARCM-M002118_seg001 | fru-Gal4 | M | 20490.05 | 0.74 |
| fru-M-500220 | 875 | FruMARCM-M002139_seg001 | fru-Gal4 | M | 20977.44 | 0.77 |
| fru-M-400082 | 1457 | FruMARCM-M001022_seg001 | fru-Gal4 | M | 19137.76 | 0.72 |
| fru-M-700007 | 2431 | FruMARCM-M000150_seg001 | fru-Gal4 | M | 16384.47 | 0.61 |
| fru-M-700008 | 2432 | FruMARCM-M000150_seg002 | fru-Gal4 | M | 16556.83 | 0.62 |
| Trh-M-000030 | 2656 | TPHMARCM-M001424_seg001 | Trh-Gal4 | M | 16768.16 | 0.61 |
| fru-F-900026 | 4765 | FruMARCM-F002084_seg001 | fru-Gal4 | F | 20248.24 | 0.75 |
| fru-F-000109 | 4770 | FruMARCM-F002089_seg001 | fru-Gal4 | F | 20651.48 | 0.76 |
| Cha-F-000205 | 4998 | ChaMARCM-F001092_seg001 | Cha-Gal4 | F | 11991.31 | 0.50 |
| Cha-F-700145 | 5456 | ChaMARCM-F000793_seg001 | Cha-Gal4 | F | 14189.86 | 0.49 |
| fru-F-200096 | 6331 | FruMARCM-F001483_seg001 | fru-Gal4 | F | 19366.79 | 0.72 |
| Trh-F-000035 | 14280 | TPHMARCM-825F_seg2 | Trh-Gal4 | F | 17084.06 | 0.64 |