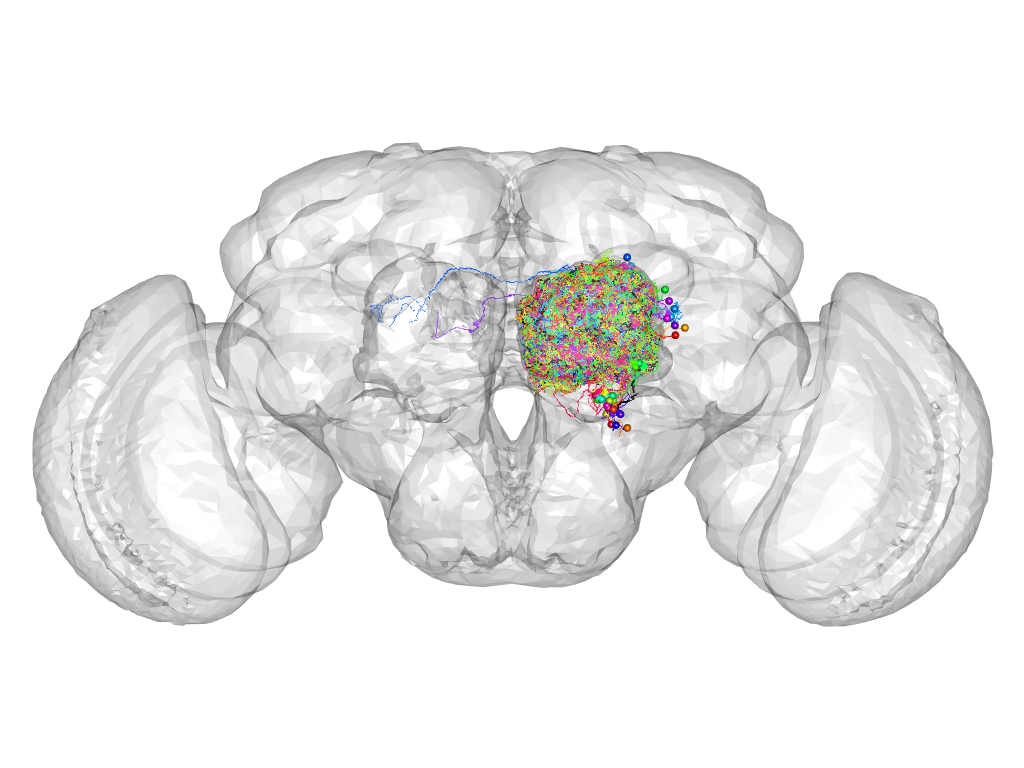
Cluster 172
Part of supercluster XII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-100092), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 806 | Trh-F-100085 | XII | 0.80 |
| 144 | Trh-M-400113 | XII | 0.70 |
| 662 | Tdc2-F-400026 | XIII | 0.63 |
| 201 | Trh-M-100007 | XII | 0.60 |
| 653 | Trh-F-000043 | XIX | 0.60 |
| 926 | VGlut-F-500134 | XII | 0.58 |
| 400 | VGlut-F-500072 | XII | 0.51 |
Cluster composition
This cluster has 29 neurons. The exemplar of this cluster (Trh-M-100092) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-100092 | 2715 | TPHMARCM-M001479_seg001 | Trh-Gal4 | M | 90111.59 | 1.00 |
| Trh-M-400121 | 2105 | TPHMARCM-M001604_seg001 | Trh-Gal4 | M | 64881.47 | 0.72 |
| Trh-M-100101 | 2172 | TPHMARCM-M001662_seg001 | Trh-Gal4 | M | 70044.28 | 0.78 |
| Trh-M-100102 | 2173 | TPHMARCM-M001663_seg001 | Trh-Gal4 | M | 70397.77 | 0.78 |
| Trh-M-900056 | 2177 | TPHMARCM-M001665_seg001 | Trh-Gal4 | M | 64170.91 | 0.72 |
| Trh-M-200099 | 2180 | TPHMARCM-M001669_seg001 | Trh-Gal4 | M | 69841.32 | 0.78 |
| Trh-M-200101 | 2182 | TPHMARCM-M001671_seg001 | Trh-Gal4 | M | 70943.63 | 0.78 |
| Trh-M-100079 | 2640 | TPHMARCM-M001409_seg001 | Trh-Gal4 | M | 69804.33 | 0.78 |
| Trh-M-200068 | 2642 | TPHMARCM-M001411_seg001 | Trh-Gal4 | M | 68934.19 | 0.78 |
| Trh-M-100089 | 2711 | TPHMARCM-M001475_seg001 | Trh-Gal4 | M | 70618.65 | 0.78 |
| Trh-M-100091 | 2713 | TPHMARCM-M001477_seg001 | Trh-Gal4 | M | 70062.64 | 0.76 |
| Trh-M-400054 | 2810 | TPHMARCM-1040M_seg1 | Trh-Gal4 | M | 64607.23 | 0.73 |
| Trh-M-100063 | 2896 | TPHMARCM-1135M_seg1 | Trh-Gal4 | M | 70015.62 | 0.78 |
| Trh-M-100067 | 2900 | TPHMARCM-1139M_seg1 | Trh-Gal4 | M | 69878.28 | 0.78 |
| Trh-M-500019 | 3552 | TPHMARCM-483M_seg1 | Trh-Gal4 | M | 66369.05 | 0.75 |
| Cha-F-500147 | 5336 | ChaMARCM-F000710_seg002 | Cha-Gal4 | F | 52142.61 | 0.65 |
| Cha-F-300158 | 7137 | ChaMARCM-F000604_seg001 | Cha-Gal4 | F | 45763.25 | 0.62 |
| Cha-F-200058 | 8254 | ChaMARCM-F000242_seg001 | Cha-Gal4 | F | 45513.07 | 0.60 |
| Gad1-F-700043 | 9688 | GadMARCM-F000537_seg001 | Gad1-Gal4 | F | 43368.83 | 0.49 |
| VGlut-F-200426 | 12022 | DvGlutMARCM-F002541_seg001 | VGlut-Gal4 | F | 45704.39 | 0.61 |
| Trh-F-100074 | 12494 | TPHMARCM-1280F_seg1 | Trh-Gal4 | F | 68747.75 | 0.78 |
| Trh-F-200131 | 12571 | TPHMARCM-F001370_seg001 | Trh-Gal4 | F | 70618.76 | 0.78 |
| TH-F-100069 | 13612 | THMARCM-555F_seg2 | TH-Gal4 | F | 37489.77 | 0.47 |
| TH-F-200102 | 13614 | THMARCM-561F_seg | TH-Gal4 | F | 40522.85 | 0.57 |
| TH-F-100107 | 13752 | THMARCM-789F_seg1 | TH-Gal4 | F | 28665.37 | 0.47 |
| Trh-F-200056 | 14013 | TPHMARCM-300F_seg2 | Trh-Gal4 | F | 69340.64 | 0.78 |
| Trh-F-200084 | 14230 | TPHMARCM-682F_seg1 | Trh-Gal4 | F | 68076.93 | 0.77 |
| Trh-F-200095 | 14287 | TPHMARCM-830F_seg1 | Trh-Gal4 | F | 69953.66 | 0.77 |
| VGlut-F-300090 | 16203 | DvGlutMARCM-F583_seg1 | VGlut-Gal4 | F | 63851.06 | 0.72 |