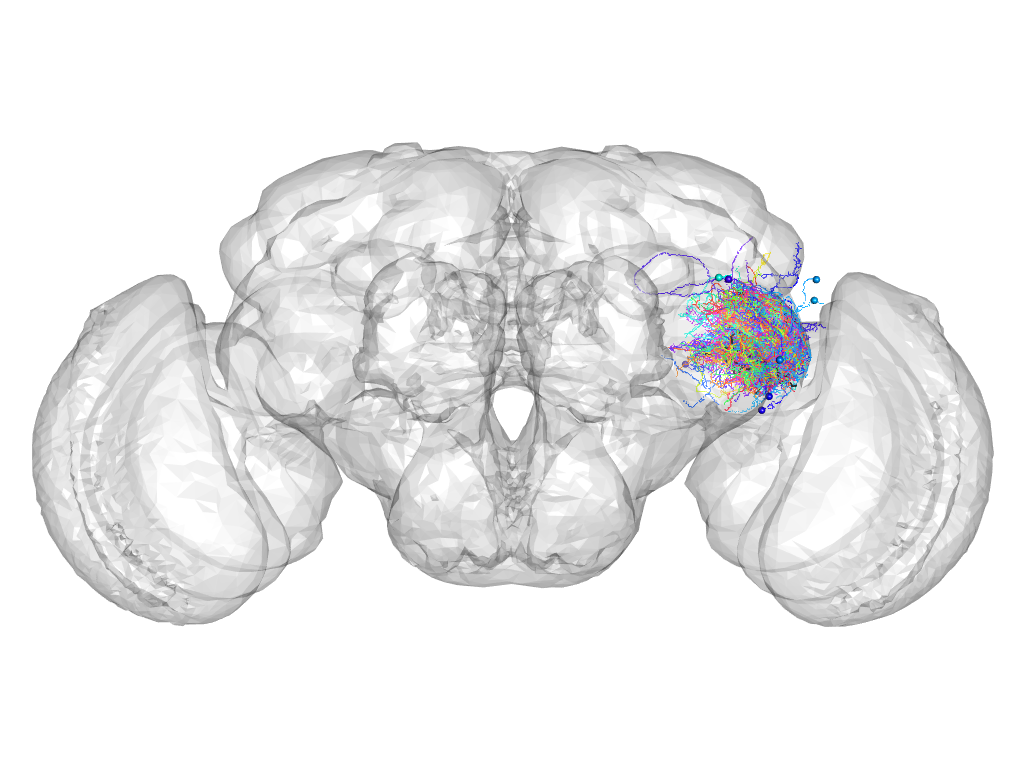
Cluster 140
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-900037), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 939 | VGlut-F-400341 | X | 0.77 |
| 321 | Cha-F-600151 | X | 0.73 |
| 895 | Trh-F-600027 | X | 0.70 |
| 876 | Trh-F-100023 | X | 0.65 |
| 171 | Trh-M-000046 | X | 0.60 |
| 962 | VGlut-F-600088 | X | 0.58 |
| 145 | Trh-M-200079 | VII | 0.55 |
Cluster composition
This cluster has 86 neurons. The exemplar of this cluster (Trh-M-900037) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-900037 | 2047 | TPHMARCM-M001554_seg001 | Trh-Gal4 | M | 32254.30 | 1.00 |
| fru-M-700151 | 79 | FruMARCM-M002279_seg001 | fru-Gal4 | M | 20716.82 | 0.69 |
| fru-M-500164 | 571 | FruMARCM-M001760_seg001 | fru-Gal4 | M | 21393.48 | 0.68 |
| fru-M-900044 | 693 | FruMARCM-M001896_seg001 | fru-Gal4 | M | 19251.21 | 0.62 |
| fru-M-200244 | 873 | FruMARCM-M002127_seg001 | fru-Gal4 | M | 20352.97 | 0.65 |
| fru-M-800043 | 984 | FruMARCM-M001164_seg001 | fru-Gal4 | M | 20266.80 | 0.64 |
| fru-M-500129 | 1063 | FruMARCM-M001240_seg001 | fru-Gal4 | M | 20795.65 | 0.67 |
| fru-M-400096 | 1473 | FruMARCM-M001032_seg001 | fru-Gal4 | M | 19905.84 | 0.67 |
| fru-M-600027 | 1697 | FruMARCM-M000850_seg001 | fru-Gal4 | M | 19188.26 | 0.62 |
| fru-M-600028 | 1698 | FruMARCM-M000850_seg002 | fru-Gal4 | M | 21604.54 | 0.69 |
| fru-M-500042 | 1968 | FruMARCM-M000615_seg001 | fru-Gal4 | M | 20791.90 | 0.69 |
| Trh-M-600059 | 1992 | TPHMARCM-1012M_seg002 | Trh-Gal4 | M | 22085.07 | 0.71 |
| Trh-M-600081 | 2042 | TPHMARCM-M001551_seg002 | Trh-Gal4 | M | 20609.29 | 0.68 |
| Trh-M-600112 | 2218 | TPHMARCM-M001709_seg001 | Trh-Gal4 | M | 20740.62 | 0.70 |
| Trh-M-200073 | 2647 | TPHMARCM-M001416_seg001 | Trh-Gal4 | M | 21042.15 | 0.68 |
| Trh-M-500108 | 2696 | TPHMARCM-M001463_seg001 | Trh-Gal4 | M | 21238.99 | 0.69 |
| Trh-M-500147 | 2752 | TPHMARCM-M001519_seg001 | Trh-Gal4 | M | 20372.02 | 0.66 |
| Trh-M-700045 | 2793 | TPHMARCM-1021M_seg1 | Trh-Gal4 | M | 20524.00 | 0.68 |
| Trh-M-700051 | 2797 | TPHMARCM-1025M_seg1 | Trh-Gal4 | M | 20357.58 | 0.63 |
| Trh-M-700052 | 2800 | TPHMARCM-1028M_seg1 | Trh-Gal4 | M | 20541.08 | 0.67 |
| Trh-M-500085 | 2827 | TPHMARCM-1055M_seg1 | Trh-Gal4 | M | 20770.09 | 0.68 |
| Trh-M-700064 | 2830 | TPHMARCM-1057M_seg1 | Trh-Gal4 | M | 21139.34 | 0.69 |
| Trh-M-600063 | 2887 | TPHMARCM-1128M_seg2 | Trh-Gal4 | M | 20564.95 | 0.68 |
| Trh-M-800010 | 2954 | TPHMARCM-895M_seg1 | Trh-Gal4 | M | 17211.14 | 0.65 |
| Trh-M-800012 | 2959 | TPHMARCM-900M_seg1 | Trh-Gal4 | M | 18931.96 | 0.63 |
| Trh-M-700014 | 2980 | TPHMARCM-918M_seg2 | Trh-Gal4 | M | 17451.98 | 0.63 |
| Trh-M-600048 | 3008 | TPHMARCM-960M_seg1 | Trh-Gal4 | M | 20856.18 | 0.71 |
| Trh-M-700018 | 3016 | TPHMARCM-968M_seg1 | Trh-Gal4 | M | 20637.82 | 0.67 |
| Trh-M-500075 | 3019 | TPHMARCM-971M_seg1 | Trh-Gal4 | M | 18243.56 | 0.67 |
| Trh-M-700021 | 3024 | TPHMARCM-976M_seg1 | Trh-Gal4 | M | 20315.87 | 0.68 |
| Trh-M-700032 | 3040 | TPHMARCM-994M_seg1 | Trh-Gal4 | M | 19014.08 | 0.63 |
| Trh-M-700036 | 3044 | TPHMARCM-998M_seg1 | Trh-Gal4 | M | 19776.20 | 0.65 |
| Trh-M-700037 | 3045 | TPHMARCM-999M_seg1 | Trh-Gal4 | M | 21249.24 | 0.68 |
| Trh-M-500016 | 3490 | TPHMARCM-362M_seg1 | Trh-Gal4 | M | 19633.39 | 0.64 |
| Trh-M-400018 | 3541 | TPHMARCM-448M_seg1 | Trh-Gal4 | M | 19314.41 | 0.64 |
| Trh-M-600008 | 3582 | TPHMARCM-520M_seg1 | Trh-Gal4 | M | 21943.60 | 0.73 |
| Trh-M-600012 | 3593 | TPHMARCM-537M_seg1 | Trh-Gal4 | M | 21201.57 | 0.70 |
| Trh-M-500036 | 3624 | TPHMARCM-598M_seg1 | Trh-Gal4 | M | 20670.55 | 0.65 |
| Trh-M-500037 | 3626 | TPHMARCM-600M_seg1 | Trh-Gal4 | M | 21595.57 | 0.69 |
| Trh-M-600028 | 3651 | TPHMARCM-665M_seg1 | Trh-Gal4 | M | 20951.13 | 0.68 |
| Trh-M-500056 | 3676 | TPHMARCM-775M_seg2 | Trh-Gal4 | M | 20489.00 | 0.65 |
| Trh-M-500059 | 3689 | TPHMARCM-804M_seg2 | Trh-Gal4 | M | 19937.79 | 0.68 |
| Cha-F-100294 | 4277 | ChaMARCM-F001353_seg001 | Cha-Gal4 | F | 20732.56 | 0.59 |
| fru-F-600093 | 4559 | FruMARCM-F001669_seg001 | fru-Gal4 | F | 17243.30 | 0.63 |
| fru-F-600099 | 4580 | FruMARCM-F001696_seg001 | fru-Gal4 | F | 21990.84 | 0.70 |
| fru-F-600110 | 4677 | FruMARCM-F001835_seg001 | fru-Gal4 | F | 20295.80 | 0.68 |
| fru-F-600116 | 4715 | FruMARCM-F002041_seg003 | fru-Gal4 | F | 19838.24 | 0.66 |
| fru-F-500246 | 4737 | FruMARCM-F002061_seg001 | fru-Gal4 | F | 21561.31 | 0.69 |
| Cha-F-300202 | 5373 | ChaMARCM-F000739_seg002 | Cha-Gal4 | F | 13788.09 | 0.51 |
| Cha-F-700148 | 5459 | ChaMARCM-F000795_seg002 | Cha-Gal4 | F | 14704.27 | 0.52 |
| Cha-F-600130 | 5594 | ChaMARCM-F000885_seg002 | Cha-Gal4 | F | 19808.69 | 0.62 |
| Cha-F-100109 | 5647 | ChaMARCM-F000923_seg001 | Cha-Gal4 | F | 19608.62 | 0.63 |
| VGlut-F-700615 | 5961 | DvGlutMARCM-F004612_seg001 | VGlut-Gal4 | F | 21351.47 | 0.71 |
| fru-F-500167 | 6234 | FruMARCM-F001197_seg001 | fru-Gal4 | F | 17909.90 | 0.55 |
| fru-F-700123 | 6236 | FruMARCM-F001216_seg001 | fru-Gal4 | F | 20434.56 | 0.66 |
| fru-F-700138 | 6300 | FruMARCM-F001397_seg002 | fru-Gal4 | F | 19674.58 | 0.66 |
| fru-F-600076 | 6305 | FruMARCM-F001425_seg001 | fru-Gal4 | F | 20686.67 | 0.68 |
| Cha-F-800016 | 7543 | ChaMARCM-F000396_seg001 | Cha-Gal4 | F | 18469.68 | 0.62 |
| Cha-F-700112 | 7586 | ChaMARCM-F000441_seg002 | Cha-Gal4 | F | 17014.78 | 0.49 |
| Cha-F-600058 | 7604 | ChaMARCM-F000451_seg002 | Cha-Gal4 | F | 18476.78 | 0.61 |
| Cha-F-700133 | 7746 | ChaMARCM-F000549_seg002 | Cha-Gal4 | F | 17766.55 | 0.53 |
| Cha-F-500070 | 8290 | ChaMARCM-F000264_seg001 | Cha-Gal4 | F | 16705.14 | 0.48 |
| VGlut-F-600577 | 9251 | DvGlutMARCM-F003312_seg001 | VGlut-Gal4 | F | 20427.55 | 0.68 |
| Gad1-F-600080 | 9695 | GadMARCM-F000542_seg003 | Gad1-Gal4 | F | 21460.55 | 0.61 |
| Trh-F-600062 | 10439 | TPHMARCM-874F_seg3 | Trh-Gal4 | F | 19416.66 | 0.67 |
| Trh-F-600063 | 10440 | TPHMARCM-874F_seg4 | Trh-Gal4 | F | 19593.00 | 0.67 |
| Gad1-F-300037 | 10565 | GadMARCM-F000088_seg001 | Gad1-Gal4 | F | 18721.55 | 0.61 |
| VGlut-F-800079 | 10789 | DvGlutMARCM-F002854_seg001 | VGlut-Gal4 | F | 19699.45 | 0.64 |
| fru-F-600002 | 11456 | FruMARCM-F000072_seg001 | fru-Gal4 | F | 18977.66 | 0.58 |
| Trh-F-600075 | 12401 | TPHMARCM-1197F_seg2 | Trh-Gal4 | F | 20085.08 | 0.67 |
| Trh-F-600086 | 12435 | TPHMARCM-1229F_seg2 | Trh-Gal4 | F | 20843.54 | 0.69 |
| Trh-F-600096 | 12449 | TPHMARCM-1243F_seg1 | Trh-Gal4 | F | 20844.40 | 0.69 |
| Trh-F-500149 | 12486 | TPHMARCM-1274F_seg2 | Trh-Gal4 | F | 21118.64 | 0.69 |
| Trh-F-500156 | 12500 | TPHMARCM-1284F_seg1 | Trh-Gal4 | F | 20043.68 | 0.68 |
| Trh-F-500161 | 12505 | TPHMARCM-1287F_seg1 | Trh-Gal4 | F | 20738.84 | 0.68 |
| Trh-F-500187 | 12545 | TPHMARCM-1333F_seg1 | Trh-Gal4 | F | 21031.31 | 0.69 |
| Trh-F-900018 | 13262 | TPHMARCM-750F_seg1 | Trh-Gal4 | F | 18464.92 | 0.65 |
| Trh-F-700025 | 13265 | TPHMARCM-753F_seg1 | Trh-Gal4 | F | 19541.29 | 0.65 |
| Trh-F-500013 | 13860 | TPHMARCM-129F_seg2 | Trh-Gal4 | F | 20485.27 | 0.66 |
| Trh-F-600024 | 14151 | TPHMARCM-571F_seg3 | Trh-Gal4 | F | 20796.86 | 0.67 |
| Trh-F-600028 | 14168 | TPHMARCM-595F_seg1 | Trh-Gal4 | F | 18542.32 | 0.58 |
| Trh-F-300084 | 14172 | TPHMARCM-610F_seg1 | Trh-Gal4 | F | 20299.70 | 0.66 |
| Trh-F-400058 | 14176 | TPHMARCM-613F_seg1 | Trh-Gal4 | F | 19760.60 | 0.65 |
| Trh-F-500102 | 14258 | TPHMARCM-802F_seg1 | Trh-Gal4 | F | 19958.16 | 0.65 |
| Trh-F-700043 | 14297 | TPHMARCM-837F_seg1 | Trh-Gal4 | F | 20255.16 | 0.66 |
| VGlut-F-600000 | 15804 | DvGlutMARCM-F214_seg1 | VGlut-Gal4 | F | 20607.20 | 0.68 |