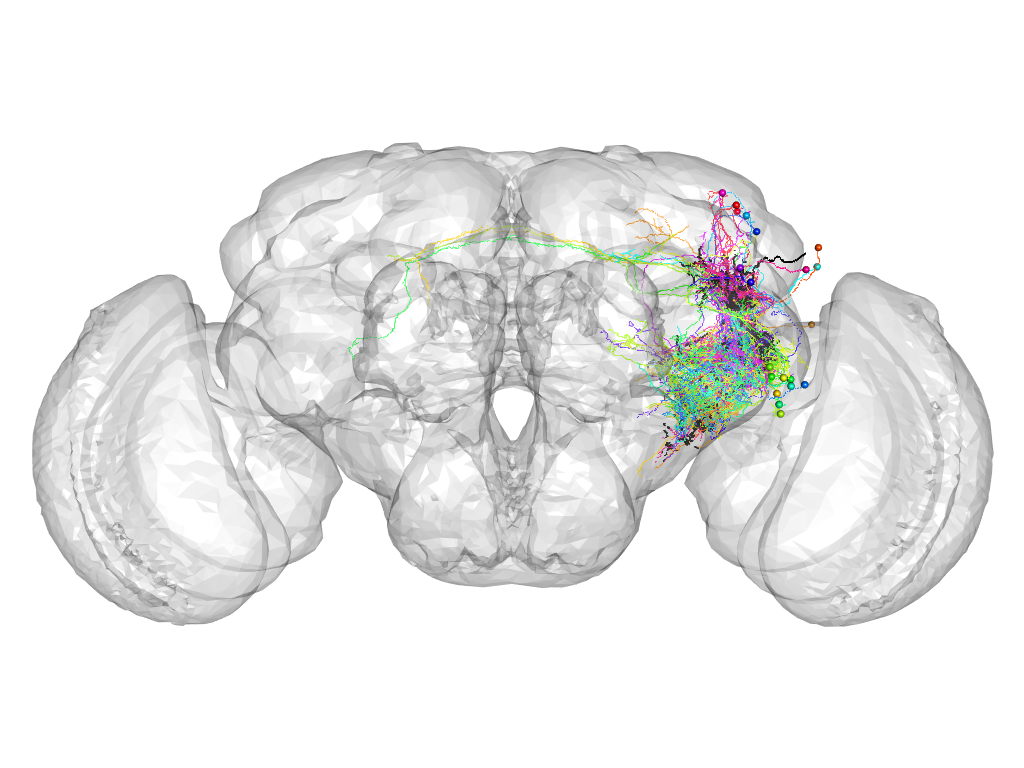
Cluster 962
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-600088), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 939 | VGlut-F-400341 | X | 0.72 |
| 200 | TH-M-000060 | X | 0.64 |
| 876 | Trh-F-100023 | X | 0.56 |
| 548 | VGlut-F-600515 | X | 0.55 |
| 865 | TH-F-000025 | X | 0.53 |
| 251 | Gad1-F-800086 | X | 0.53 |
| 321 | Cha-F-600151 | X | 0.52 |
Cluster composition
This cluster has 23 neurons. The exemplar of this cluster (VGlut-F-600088) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-600088 | 15364 | DvGlutMARCM-F1716_seg1 | VGlut-Gal4 | F | 115202.06 | 1.00 |
| fru-M-000040 | 1079 | FruMARCM-M001255_seg001 | fru-Gal4 | M | 65503.48 | 0.60 |
| Trh-M-500128 | 1314 | TPHMARCM-M001500_seg001 | Trh-Gal4 | M | 48804.40 | 0.54 |
| Cha-F-700186 | 3877 | ChaMARCM-F001543_seg002 | Cha-Gal4 | F | 45211.70 | 0.39 |
| Gad1-F-700050 | 4077 | GadMARCM-F000667_seg001 | Gad1-Gal4 | F | 76157.17 | 0.57 |
| Cha-F-100253 | 4166 | ChaMARCM-F001277_seg001 | Cha-Gal4 | F | 77283.94 | 0.66 |
| Cha-F-000305 | 4223 | ChaMARCM-F001312_seg001 | Cha-Gal4 | F | 71259.53 | 0.53 |
| Cha-F-200333 | 4300 | ChaMARCM-F001367_seg003 | Cha-Gal4 | F | 69453.57 | 0.62 |
| Cha-F-200355 | 4347 | ChaMARCM-F001404_seg001 | Cha-Gal4 | F | 78937.34 | 0.68 |
| Cha-F-000360 | 4419 | ChaMARCM-F001467_seg002 | Cha-Gal4 | F | 57403.02 | 0.50 |
| fru-F-400191 | 4727 | FruMARCM-F002051_seg004 | fru-Gal4 | F | 71894.52 | 0.65 |
| fru-F-600117 | 4739 | FruMARCM-F002063_seg001 | fru-Gal4 | F | 72522.90 | 0.61 |
| fru-F-400203 | 4758 | FruMARCM-F002078_seg001 | fru-Gal4 | F | 57862.65 | 0.54 |
| Cha-F-200174 | 5567 | ChaMARCM-F000866_seg001 | Cha-Gal4 | F | 64192.14 | 0.59 |
| fru-F-700125 | 6240 | FruMARCM-F001219_seg001 | fru-Gal4 | F | 73432.63 | 0.61 |
| VGlut-F-500788 | 6467 | DvGlutMARCM-F004251_seg002 | VGlut-Gal4 | F | 54692.96 | 0.57 |
| Cha-F-600093 | 7191 | ChaMARCM-F000640_seg001 | Cha-Gal4 | F | 63255.21 | 0.59 |
| Cha-F-300128 | 7554 | ChaMARCM-F000405_seg003 | Cha-Gal4 | F | 44502.55 | 0.39 |
| 5-HT1B-F-000005 | 11623 | 5HT1bMARCM-F000017_seg002 | 5-HT1B-Gal4 | F | 71248.37 | 0.63 |
| Trh-F-900008 | 13927 | TPHMARCM-217F_seg1 | Trh-Gal4 | F | 66543.25 | 0.55 |
| VGlut-F-500256 | 15423 | DvGlutMARCM-F1780_seg1 | VGlut-Gal4 | F | 65359.30 | 0.65 |
| VGlut-F-000002 | 15603 | DvGlutMARCM-F019_seg1 | VGlut-Gal4 | F | 85872.66 | 0.77 |
| VGlut-F-600014 | 15879 | DvGlutMARCM-F280_seg2 | VGlut-Gal4 | F | 70297.67 | 0.65 |