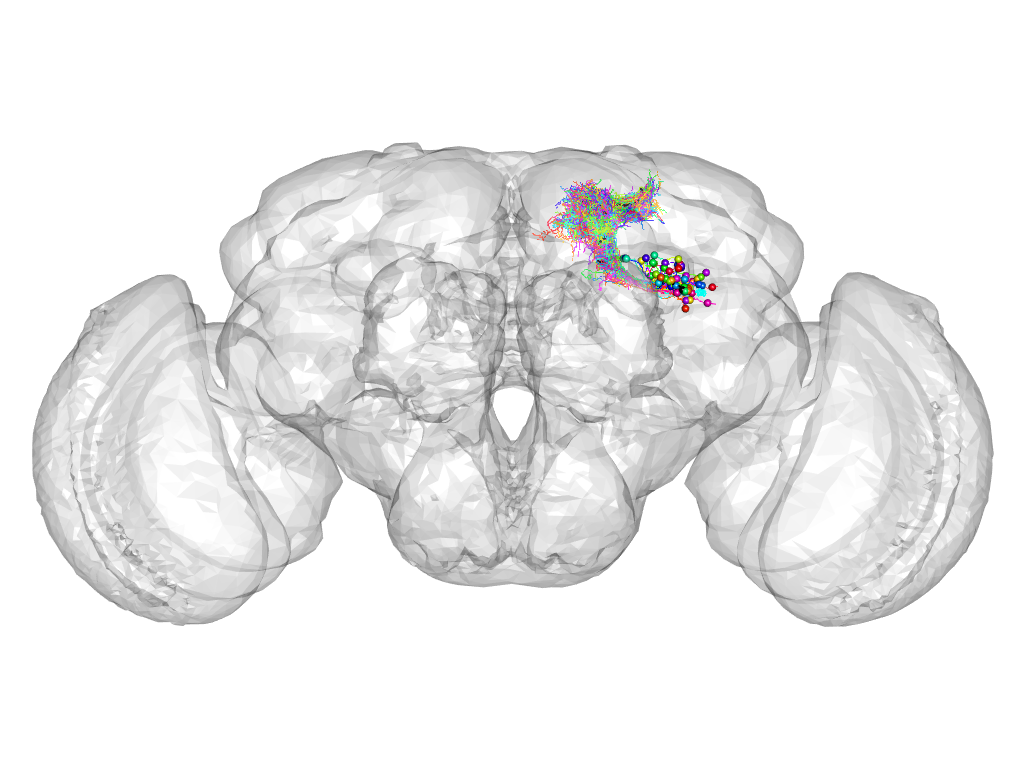
Cluster 135
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-500050), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 877 | Trh-F-000012 | XIX | 0.78 |
| 204 | Trh-M-100025 | XIV | 0.76 |
| 198 | TH-M-000032 | XIX | 0.74 |
| 190 | TH-M-200014 | XIX | 0.72 |
| 1003 | VGlut-F-000024 | XIX | 0.65 |
| 994 | VGlut-F-200045 | XIX | 0.62 |
| 136 | TH-M-000013 | XVII | 0.61 |
| 941 | VGlut-F-200251 | XIX | 0.61 |
| 884 | Trh-F-500028 | XIX | 0.61 |
| 182 | Trh-M-600037 | XIX | 0.53 |
| 867 | TH-F-100077 | XIX | 0.52 |
Cluster composition
This cluster has 56 neurons. The exemplar of this cluster (Trh-M-500050) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-500050 | 1985 | TPHMARCM-0769M_seg001 | Trh-Gal4 | M | 11434.79 | 1.00 |
| Trh-M-900045 | 2055 | TPHMARCM-M001560_seg001 | Trh-Gal4 | M | 8316.07 | 0.73 |
| Trh-M-500177 | 2082 | TPHMARCM-M001583_seg001 | Trh-Gal4 | M | 8113.84 | 0.69 |
| Trh-M-500182 | 2087 | TPHMARCM-M001588_seg001 | Trh-Gal4 | M | 8304.39 | 0.72 |
| Trh-M-500191 | 2109 | TPHMARCM-M001607_seg001 | Trh-Gal4 | M | 8034.99 | 0.70 |
| Trh-M-600096 | 2196 | TPHMARCM-M001686_seg001 | Trh-Gal4 | M | 7779.98 | 0.66 |
| Trh-M-500154 | 2546 | TPHMARCM-M001525_seg001 | Trh-Gal4 | M | 8235.38 | 0.72 |
| Trh-M-500164 | 2555 | TPHMARCM-M001531_seg001 | Trh-Gal4 | M | 8552.07 | 0.74 |
| Trh-M-700072 | 2594 | TPHMARCM-1184M_seg1 | Trh-Gal4 | M | 7687.88 | 0.69 |
| Trh-M-500095 | 2662 | TPHMARCM-M001427_seg002 | Trh-Gal4 | M | 8262.44 | 0.73 |
| Trh-M-500104 | 2683 | TPHMARCM-M001449_seg001 | Trh-Gal4 | M | 8162.75 | 0.71 |
| Trh-M-500116 | 2704 | TPHMARCM-M001467_seg001 | Trh-Gal4 | M | 8590.72 | 0.75 |
| Trh-M-500140 | 2737 | TPHMARCM-M001503_seg002 | Trh-Gal4 | M | 8237.05 | 0.73 |
| Trh-M-700059 | 2808 | TPHMARCM-1038M_seg1 | Trh-Gal4 | M | 7992.77 | 0.72 |
| Trh-M-400061 | 2826 | TPHMARCM-1054M_seg2 | Trh-Gal4 | M | 7828.87 | 0.71 |
| Trh-M-400072 | 2843 | TPHMARCM-1069M_seg1 | Trh-Gal4 | M | 8187.90 | 0.71 |
| Trh-M-400099 | 2877 | TPHMARCM-1122M_seg1 | Trh-Gal4 | M | 8226.91 | 0.71 |
| Trh-M-500065 | 2953 | TPHMARCM-894M_seg1 | Trh-Gal4 | M | 8142.74 | 0.72 |
| Trh-M-800014 | 2961 | TPHMARCM-902M_seg1 | Trh-Gal4 | M | 8114.03 | 0.72 |
| Trh-M-800015 | 2962 | TPHMARCM-902M_seg2 | Trh-Gal4 | M | 8397.14 | 0.74 |
| Trh-M-900021 | 2963 | TPHMARCM-903M_seg1 | Trh-Gal4 | M | 8389.95 | 0.75 |
| Trh-M-900024 | 3012 | TPHMARCM-964M_seg1 | Trh-Gal4 | M | 7646.23 | 0.68 |
| Trh-M-900025 | 3013 | TPHMARCM-965M_seg1 | Trh-Gal4 | M | 8400.84 | 0.74 |
| Trh-M-900026 | 3015 | TPHMARCM-967M_seg1 | Trh-Gal4 | M | 8588.93 | 0.75 |
| Trh-M-700029 | 3037 | TPHMARCM-990M_seg1 | Trh-Gal4 | M | 8344.73 | 0.72 |
| Trh-M-700004 | 3655 | TPHMARCM-670M_seg1 | Trh-Gal4 | M | 8384.14 | 0.73 |
| Trh-M-700008 | 3680 | TPHMARCM-794M_seg1 | Trh-Gal4 | M | 8061.80 | 0.71 |
| Trh-M-500057 | 3683 | TPHMARCM-795M_seg1 | Trh-Gal4 | M | 8466.27 | 0.75 |
| Trh-F-500130 | 10435 | TPHMARCM-1255F_seg1 | Trh-Gal4 | F | 8400.52 | 0.70 |
| Trh-F-500223 | 11211 | TPHMARCM-F001653_seg001 | Trh-Gal4 | F | 8507.66 | 0.74 |
| VGlut-F-500308 | 12329 | DvGlutMARCM-F1931_seg1 | VGlut-Gal4 | F | 8293.54 | 0.72 |
| Trh-F-500107 | 12390 | TPHMARCM-1188F_seg1 | Trh-Gal4 | F | 8778.20 | 0.76 |
| Trh-F-700055 | 12393 | TPHMARCM-1191F_seg1 | Trh-Gal4 | F | 8308.79 | 0.74 |
| Trh-F-500109 | 12404 | TPHMARCM-1199F_seg1 | Trh-Gal4 | F | 7974.73 | 0.68 |
| Trh-F-500110 | 12405 | TPHMARCM-1200F_seg1 | Trh-Gal4 | F | 8361.68 | 0.73 |
| Trh-F-500116 | 12411 | TPHMARCM-1204F_seg1 | Trh-Gal4 | F | 7910.45 | 0.71 |
| Trh-F-700060 | 12425 | TPHMARCM-1217F_seg1 | Trh-Gal4 | F | 8013.84 | 0.71 |
| Trh-F-600100 | 12453 | TPHMARCM-1245F_seg1 | Trh-Gal4 | F | 7792.66 | 0.70 |
| Trh-F-500123 | 12455 | TPHMARCM-1247F_seg1 | Trh-Gal4 | F | 8415.93 | 0.73 |
| Trh-F-500136 | 12471 | TPHMARCM-1260F_seg001 | Trh-Gal4 | F | 7908.37 | 0.69 |
| Trh-F-500147 | 12484 | TPHMARCM-1273F_seg1 | Trh-Gal4 | F | 8444.04 | 0.73 |
| Trh-F-500159 | 12503 | TPHMARCM-1286F_seg1 | Trh-Gal4 | F | 8156.31 | 0.70 |
| Trh-F-500164 | 12508 | TPHMARCM-1290F_seg1 | Trh-Gal4 | F | 8092.36 | 0.70 |
| Trh-F-500166 | 12510 | TPHMARCM-1292F_seg1 | Trh-Gal4 | F | 8130.18 | 0.71 |
| Trh-F-500179 | 12537 | TPHMARCM-1320F_seg1 | Trh-Gal4 | F | 8005.80 | 0.71 |
| Trh-F-700046 | 12716 | TPHMARCM-879F_seg1 | Trh-Gal4 | F | 8383.86 | 0.72 |
| Trh-F-500079 | 13220 | TPHMARCM-702F_seg1 | Trh-Gal4 | F | 8214.69 | 0.72 |
| Trh-F-400071 | 13227 | TPHMARCM-714F_seg1 | Trh-Gal4 | F | 7818.99 | 0.67 |
| Trh-F-500000 | 13847 | TPHMARCM-119F_seg1 | Trh-Gal4 | F | 8177.47 | 0.72 |
| Trh-F-500030 | 14025 | TPHMARCM-311F_seg2 | Trh-Gal4 | F | 8136.89 | 0.70 |
| Trh-F-500045 | 14126 | TPHMARCM-513F_seg1 | Trh-Gal4 | F | 8321.02 | 0.75 |
| Trh-F-200070 | 14137 | TPHMARCM-531F_seg1 | Trh-Gal4 | F | 8576.83 | 0.73 |
| Trh-F-200071 | 14138 | TPHMARCM-531F_seg2 | Trh-Gal4 | F | 8026.30 | 0.69 |
| Trh-F-400053 | 14141 | TPHMARCM-553F_seg1 | Trh-Gal4 | F | 8233.52 | 0.73 |
| Trh-F-600036 | 14221 | TPHMARCM-668F_seg1 | Trh-Gal4 | F | 8514.10 | 0.74 |
| Trh-F-500069 | 14222 | TPHMARCM-674F_seg1 | Trh-Gal4 | F | 8129.23 | 0.69 |