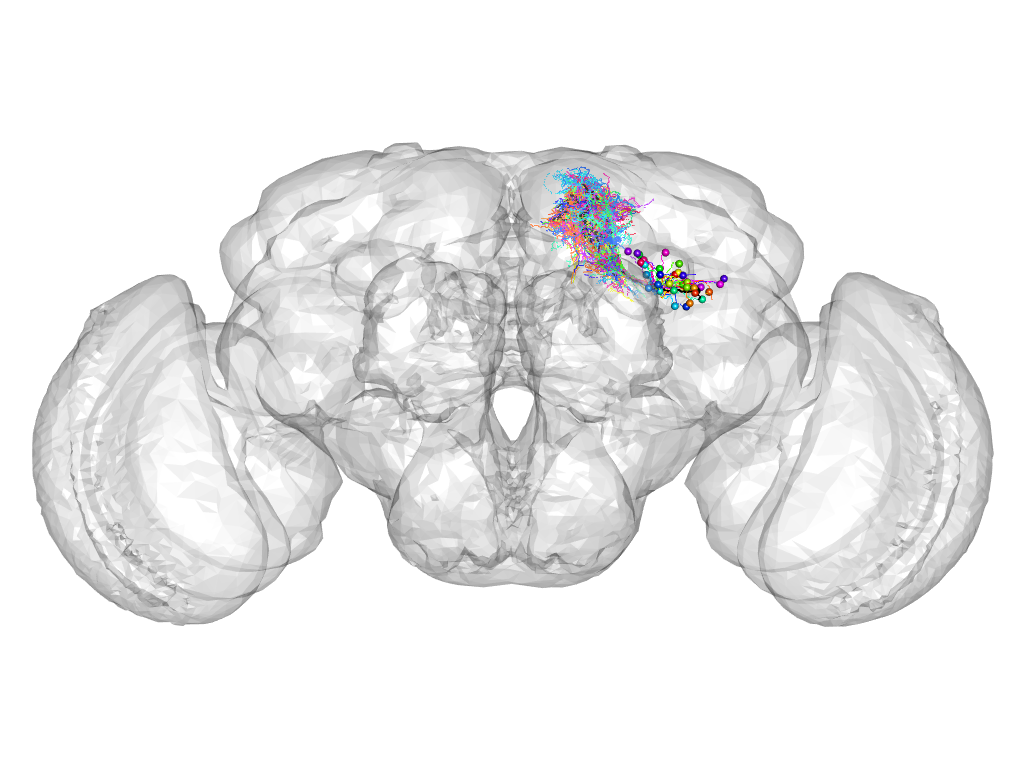
Cluster 884
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-500028), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 204 | Trh-M-100025 | XIV | 0.78 |
| 877 | Trh-F-000012 | XIX | 0.76 |
| 198 | TH-M-000032 | XIX | 0.75 |
| 994 | VGlut-F-200045 | XIX | 0.71 |
| 182 | Trh-M-600037 | XIX | 0.70 |
| 135 | Trh-M-500050 | XIX | 0.68 |
| 190 | TH-M-200014 | XIX | 0.68 |
| 867 | TH-F-100077 | XIX | 0.64 |
| 1003 | VGlut-F-000024 | XIX | 0.64 |
| 1032 | VGlut-F-000077 | XIX | 0.62 |
| 186 | TH-M-600001 | XIX | 0.61 |
| 197 | TH-M-200095 | XIX | 0.60 |
| 42 | fru-M-300180 | XIV | 0.59 |
| 941 | VGlut-F-200251 | XIX | 0.59 |
| 80 | fru-M-400195 | XIV | 0.58 |
| 136 | TH-M-000013 | XVII | 0.57 |
| 143 | Trh-M-400111 | XIX | 0.56 |
| 564 | VGlut-F-000535 | XIX | 0.51 |
| 848 | TH-F-700007 | XIX | 0.51 |
| 871 | TH-F-600003 | XIX | 0.51 |
Cluster composition
This cluster has 37 neurons. The exemplar of this cluster (Trh-F-500028) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-500028 | 14021 | TPHMARCM-308F_seg1 | Trh-Gal4 | F | 19965.32 | 1.00 |
| fru-M-400125 | 1050 | FruMARCM-M001230_seg001 | fru-Gal4 | M | 14554.55 | 0.76 |
| Trh-M-600085 | 2098 | TPHMARCM-M001597_seg001 | Trh-Gal4 | M | 14974.41 | 0.76 |
| Trh-M-500156 | 2548 | TPHMARCM-M001527_seg001 | Trh-Gal4 | M | 12116.70 | 0.63 |
| Trh-M-500171 | 2560 | TPHMARCM-M001537_seg001 | Trh-Gal4 | M | 14798.55 | 0.74 |
| Trh-M-500090 | 2657 | TPHMARCM-M001425_seg001 | Trh-Gal4 | M | 14674.30 | 0.75 |
| Trh-M-500111 | 2699 | TPHMARCM-M001464_seg002 | Trh-Gal4 | M | 14376.51 | 0.74 |
| Trh-M-500141 | 2738 | TPHMARCM-M001504_seg001 | Trh-Gal4 | M | 14279.61 | 0.70 |
| Trh-M-700063 | 2829 | TPHMARCM-1056M_seg2 | Trh-Gal4 | M | 14376.97 | 0.74 |
| Trh-M-400075 | 2846 | TPHMARCM-1071M_seg2 | Trh-Gal4 | M | 14280.20 | 0.73 |
| Trh-M-400041 | 2927 | TPHMARCM-862M_seg2 | Trh-Gal4 | M | 13293.46 | 0.70 |
| Trh-M-500067 | 2964 | TPHMARCM-904M_seg1 | Trh-Gal4 | M | 14343.51 | 0.74 |
| Trh-M-500017 | 3491 | TPHMARCM-363M_seg1 | Trh-Gal4 | M | 14102.76 | 0.73 |
| Trh-M-500028 | 3580 | TPHMARCM-517M_seg1 | Trh-Gal4 | M | 14058.66 | 0.74 |
| Trh-M-500040 | 3629 | TPHMARCM-602M_seg2 | Trh-Gal4 | M | 14167.67 | 0.74 |
| VGlut-F-500867 | 5797 | DvGlutMARCM-F004496_seg001 | VGlut-Gal4 | F | 13905.04 | 0.72 |
| VGlut-F-700606 | 5947 | DvGlutMARCM-F004601_seg001 | VGlut-Gal4 | F | 11843.53 | 0.64 |
| Trh-F-700002 | 5986 | TPHMARCM-322F_seg1 | Trh-Gal4 | F | 14547.14 | 0.74 |
| Gad1-F-700049 | 6408 | GadMARCM-F000566_seg001 | Gad1-Gal4 | F | 14256.83 | 0.73 |
| VGlut-F-500734 | 6707 | DvGlutMARCM-F003921_seg001 | VGlut-Gal4 | F | 12130.85 | 0.63 |
| VGlut-F-900118 | 6750 | DvGlutMARCM-F003955_seg001 | VGlut-Gal4 | F | 12180.90 | 0.58 |
| VGlut-F-600717 | 6789 | DvGlutMARCM-F003982_seg001 | VGlut-Gal4 | F | 13148.23 | 0.67 |
| VGlut-F-500759 | 6820 | DvGlutMARCM-F004004_seg001 | VGlut-Gal4 | F | 12428.62 | 0.62 |
| VGlut-F-800062 | 12113 | DvGlutMARCM-F002610_seg001 | VGlut-Gal4 | F | 14501.55 | 0.69 |
| Trh-F-700050 | 12383 | TPHMARCM-1179F_seg1 | Trh-Gal4 | F | 14374.29 | 0.74 |
| Trh-F-500125 | 12457 | TPHMARCM-1248F_seg1 | Trh-Gal4 | F | 14695.04 | 0.74 |
| Trh-F-500129 | 12461 | TPHMARCM-1249F_seg1 | Trh-Gal4 | F | 14266.87 | 0.72 |
| Trh-F-500158 | 12502 | TPHMARCM-1285F_seg2 | Trh-Gal4 | F | 14318.78 | 0.74 |
| Trh-F-500163 | 12507 | TPHMARCM-1288F_seg2 | Trh-Gal4 | F | 13572.32 | 0.71 |
| Trh-F-500181 | 12539 | TPHMARCM-1321F_seg2 | Trh-Gal4 | F | 14347.61 | 0.70 |
| Trh-F-500087 | 13268 | TPHMARCM-756F_seg1 | Trh-Gal4 | F | 14936.24 | 0.73 |
| Trh-F-400040 | 13997 | TPHMARCM-287F_seg1 | Trh-Gal4 | F | 14794.19 | 0.74 |
| Trh-F-600019 | 14133 | TPHMARCM-523F_seg1 | Trh-Gal4 | F | 14405.25 | 0.72 |
| Trh-F-600033 | 14207 | TPHMARCM-648F_seg1 | Trh-Gal4 | F | 14967.15 | 0.72 |
| Trh-F-700031 | 14266 | TPHMARCM-814F_seg1 | Trh-Gal4 | F | 14090.94 | 0.72 |
| Trh-F-700033 | 14269 | TPHMARCM-817F_seg1 | Trh-Gal4 | F | 14016.47 | 0.73 |
| Trh-F-900027 | 14276 | TPHMARCM-822F_seg2 | Trh-Gal4 | F | 13718.17 | 0.68 |