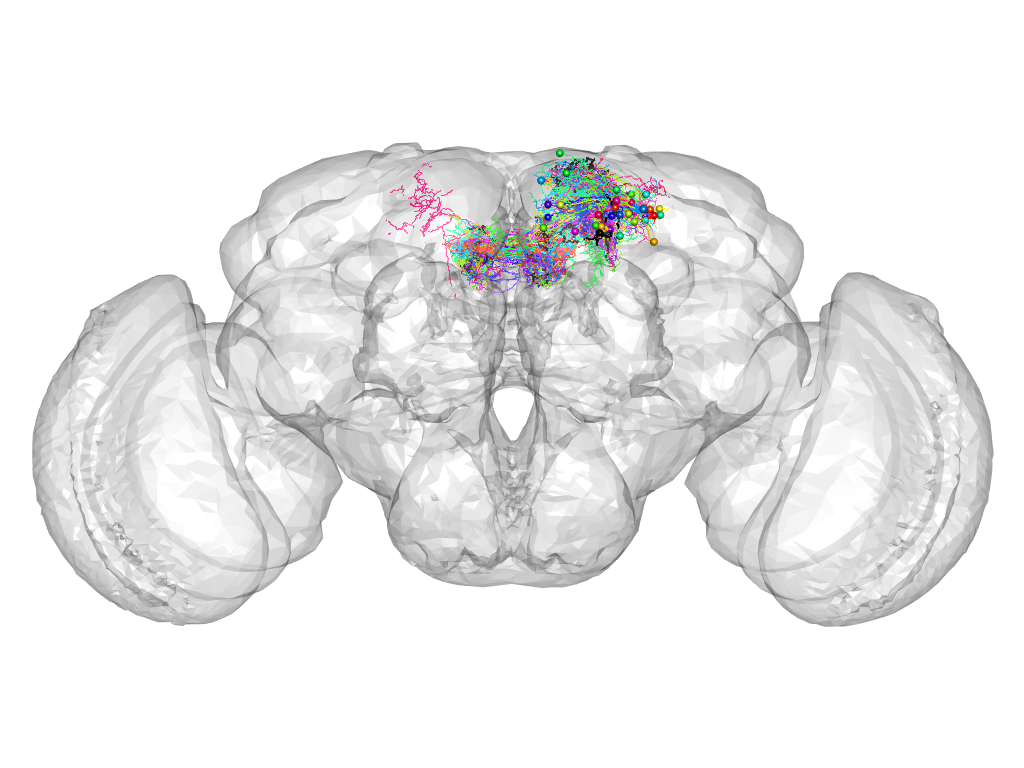
Cluster 867
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-F-100077), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 186 | TH-M-600001 | XIX | 0.55 |
| 204 | Trh-M-100025 | XIV | 0.48 |
| 877 | Trh-F-000012 | XIX | 0.47 |
| 871 | TH-F-600003 | XIX | 0.46 |
| 197 | TH-M-200095 | XIX | 0.45 |
Cluster composition
This cluster has 39 neurons. The exemplar of this cluster (TH-F-100077) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-F-100077 | 13644 | THMARCM-648F_seg | TH-Gal4 | F | 37037.78 | 1.00 |
| TH-M-700004 | 1724 | THMARCM_117M_seg1 | TH-Gal4 | M | 18508.82 | 0.57 |
| Trh-M-500194 | 2114 | TPHMARCM-M001612_seg001 | Trh-Gal4 | M | 18438.68 | 0.58 |
| Trh-M-600098 | 2198 | TPHMARCM-M001688_seg001 | Trh-Gal4 | M | 17916.77 | 0.59 |
| Trh-M-600104 | 2207 | TPHMARCM-M001695_seg002 | Trh-Gal4 | M | 18684.02 | 0.60 |
| TH-M-700012 | 3047 | THMARCM-128M_seg1 | TH-Gal4 | M | 19404.88 | 0.57 |
| TH-M-500004 | 3060 | THMARCM-618M_seg | TH-Gal4 | M | 20961.32 | 0.63 |
| TH-M-600008 | 3067 | THMARCM-653M_seg | TH-Gal4 | M | 19418.72 | 0.62 |
| TH-M-700002 | 3080 | THMARCM-048M_seg1 | TH-Gal4 | M | 17254.64 | 0.55 |
| TH-M-700008 | 3111 | THMARCM-125M_seg1 | TH-Gal4 | M | 21329.20 | 0.66 |
| TH-M-700016 | 3117 | THMARCM-145M_seg1 | TH-Gal4 | M | 21037.91 | 0.63 |
| TH-M-700017 | 3118 | THMARCM-147M_seg1 | TH-Gal4 | M | 17208.63 | 0.55 |
| TH-M-600007 | 3353 | THMARCM-685M_seg | TH-Gal4 | M | 17263.35 | 0.54 |
| Gad1-F-000098 | 4110 | GadMARCM-F000695_seg001 | Gad1-Gal4 | F | 14805.75 | 0.51 |
| Cha-F-100305 | 4329 | ChaMARCM-F001390_seg001 | Cha-Gal4 | F | 16572.60 | 0.53 |
| Cha-F-300183 | 5271 | ChaMARCM-F000667_seg002 | Cha-Gal4 | F | 15927.29 | 0.50 |
| VGlut-F-600727 | 6841 | DvGlutMARCM-F004022_seg001 | VGlut-Gal4 | F | 20042.51 | 0.61 |
| fru-F-400105 | 7872 | FruMARCM-F000796_seg002 | fru-Gal4 | F | 19552.54 | 0.55 |
| fru-F-500042 | 9956 | FruMARCM-F000548_seg001 | fru-Gal4 | F | 19519.17 | 0.56 |
| Gad1-F-900030 | 11302 | GadMARCM-F000275_seg001 | Gad1-Gal4 | F | 17201.99 | 0.57 |
| Trh-F-700062 | 12429 | TPHMARCM-1225F_seg1 | Trh-Gal4 | F | 17188.77 | 0.60 |
| Trh-F-500194 | 12562 | TPHMARCM-F001350_seg001 | Trh-Gal4 | F | 18184.47 | 0.58 |
| Trh-F-400069 | 13225 | TPHMARCM-712F_seg1 | Trh-Gal4 | F | 18062.24 | 0.56 |
| TH-F-700001 | 13325 | THMARCM-046F_seg1 | TH-Gal4 | F | 20424.57 | 0.59 |
| TH-F-700003 | 13327 | THMARCM-049F_seg1 | TH-Gal4 | F | 19636.50 | 0.63 |
| TH-F-700005 | 13330 | THMARCM-055F_seg1 | TH-Gal4 | F | 18791.80 | 0.55 |
| TH-F-700006 | 13331 | THMARCM-056F_seg1 | TH-Gal4 | F | 18112.23 | 0.56 |
| TH-F-700018 | 13364 | THMARCM-108_seg1 | TH-Gal4 | F | 15763.95 | 0.53 |
| TH-F-700025 | 13371 | THMARCM-116_seg1 | TH-Gal4 | F | 17914.20 | 0.59 |
| TH-F-700026 | 13372 | THMARCM-120F_seg1 | TH-Gal4 | F | 12435.99 | 0.44 |
| TH-F-700027 | 13373 | THMARCM-130F_seg1 | TH-Gal4 | F | 20946.45 | 0.65 |
| TH-F-700032 | 13378 | THMARCM-138F_seg1 | TH-Gal4 | F | 13109.12 | 0.51 |
| TH-F-700034 | 13380 | THMARCM-140F_seg1 | TH-Gal4 | F | 18567.83 | 0.62 |
| TH-F-700042 | 13654 | THMARCM-666F_seg | TH-Gal4 | F | 18709.41 | 0.58 |
| TH-F-500003 | 13665 | THMARCM-679F_seg | TH-Gal4 | F | 18248.26 | 0.55 |
| TH-F-500004 | 13666 | THMARCM-680F_seg | TH-Gal4 | F | 18633.35 | 0.57 |
| TH-F-600001 | 13703 | THMARCM-726F_seg | TH-Gal4 | F | 23791.92 | 0.59 |
| TH-F-600002 | 13704 | THMARCM-727F_seg | TH-Gal4 | F | 17021.26 | 0.53 |
| Trh-F-400079 | 14261 | TPHMARCM-809F-01_seg1 | Trh-Gal4 | F | 18381.23 | 0.60 |