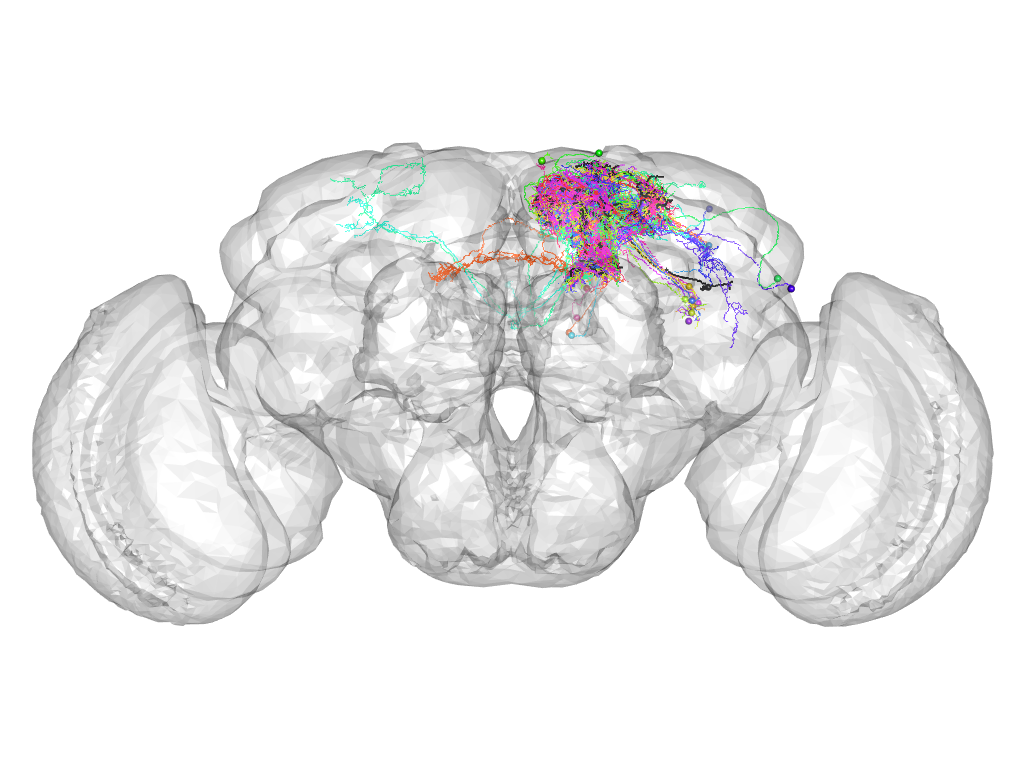
Cluster 198
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-M-000032), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 204 | Trh-M-100025 | XIV | 0.76 |
| 197 | TH-M-200095 | XIX | 0.72 |
| 877 | Trh-F-000012 | XIX | 0.69 |
| 136 | TH-M-000013 | XVII | 0.69 |
| 190 | TH-M-200014 | XIX | 0.67 |
| 1003 | VGlut-F-000024 | XIX | 0.65 |
| 564 | VGlut-F-000535 | XIX | 0.64 |
| 941 | VGlut-F-200251 | XIX | 0.63 |
| 143 | Trh-M-400111 | XIX | 0.61 |
| 994 | VGlut-F-200045 | XIX | 0.61 |
| 864 | TH-F-100064 | XIX | 0.61 |
| 1032 | VGlut-F-000077 | XIX | 0.59 |
| 867 | TH-F-100077 | XIX | 0.58 |
| 1004 | VGlut-F-100016 | VI | 0.53 |
Cluster composition
This cluster has 22 neurons. The exemplar of this cluster (TH-M-000032) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-M-000032 | 3332 | THMARCM-603M-X2_seg2 | TH-Gal4 | M | 182911.03 | 1.00 |
| fru-M-400179 | 419 | FruMARCM-M001935_seg003 | fru-Gal4 | M | 113175.94 | 0.68 |
| fru-M-100117 | 1124 | FruMARCM-M001320_seg001 | fru-Gal4 | M | 117327.27 | 0.51 |
| TH-M-000046 | 2028 | THMARCM-631M_seg01 | TH-Gal4 | M | 139627.72 | 0.78 |
| TH-M-000029 | 3056 | THMARCM-601M_seg | TH-Gal4 | M | 147282.77 | 0.81 |
| TH-M-000021 | 3323 | THMARCM-570M_seg | TH-Gal4 | M | 144727.45 | 0.80 |
| TH-M-000043 | 3341 | THMARCM-628M_seg | TH-Gal4 | M | 144735.66 | 0.80 |
| Cha-F-100354 | 3854 | ChaMARCM-F001525_seg001 | Cha-Gal4 | F | 119908.52 | 0.69 |
| Gad1-F-800073 | 4013 | GadMARCM-F000626_seg001 | Gad1-Gal4 | F | 117214.23 | 0.69 |
| Cha-F-400223 | 4838 | ChaMARCM-F000964_seg001 | Cha-Gal4 | F | 118717.88 | 0.67 |
| Cha-F-300184 | 5272 | ChaMARCM-F000668_seg001 | Cha-Gal4 | F | 107618.01 | 0.49 |
| Cha-F-300209 | 5441 | ChaMARCM-F000782_seg001 | Cha-Gal4 | F | 110708.58 | 0.53 |
| VGlut-F-100386 | 5700 | DvGlutMARCM-F004411_seg001 | VGlut-Gal4 | F | 109997.06 | 0.66 |
| Cha-F-300057 | 8200 | ChaMARCM-F000210_seg001 | Cha-Gal4 | F | 76858.11 | 0.59 |
| TH-F-000046 | 10463 | THMARCM-660F_seg001 | TH-Gal4 | F | 144336.99 | 0.80 |
| Gad1-F-200037 | 10496 | GadMARCM-F000151_seg001 | Gad1-Gal4 | F | 73659.52 | 0.48 |
| VGlut-F-200375 | 13020 | DvGlutMARCM-F2046_seg1 | VGlut-Gal4 | F | 104291.32 | 0.56 |
| TH-F-000018 | 13593 | THMARCM-526F_seg1 | TH-Gal4 | F | 146226.84 | 0.80 |
| TH-F-000019 | 13595 | THMARCM-528F_seg1 | TH-Gal4 | F | 146307.42 | 0.80 |
| TH-F-000045 | 13652 | THMARCM-659F_seg | TH-Gal4 | F | 144647.04 | 0.79 |
| VGlut-F-100090 | 14352 | DvGlutMARCM-F1036_seg1 | VGlut-Gal4 | F | 120402.51 | 0.70 |
| VGlut-F-300066 | 15958 | DvGlutMARCM-F351-X2_seg1 | VGlut-Gal4 | F | 122990.01 | 0.71 |