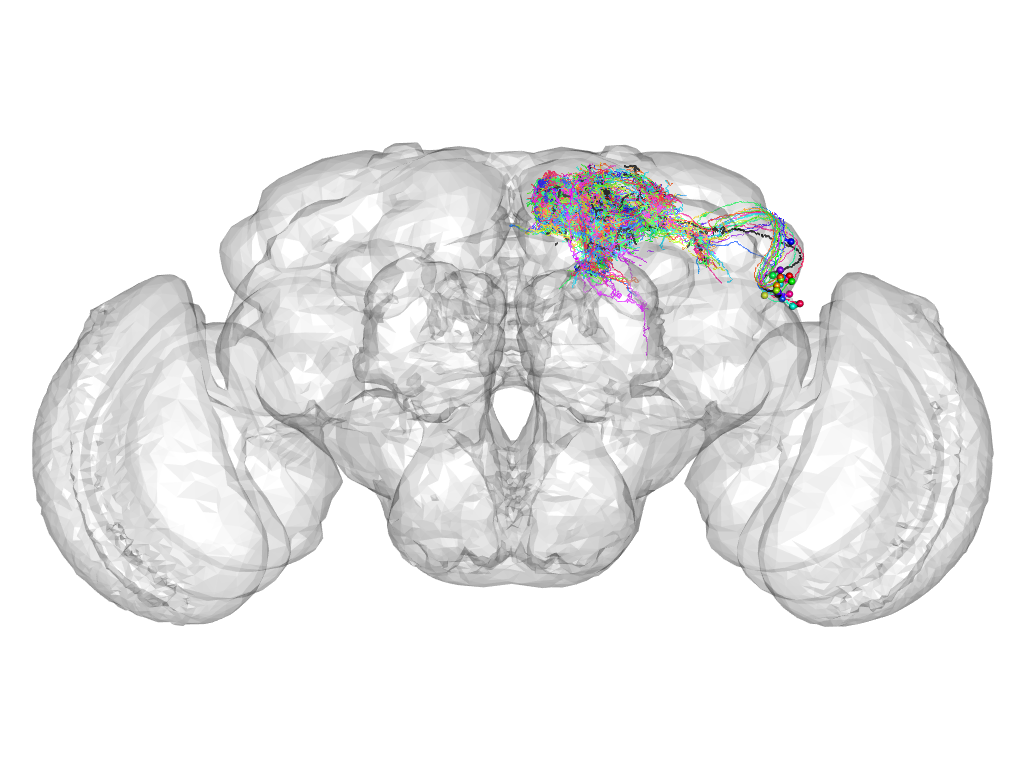
Cluster 864
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-F-100064), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 136 | TH-M-000013 | XVII | 0.70 |
| 198 | TH-M-000032 | XIX | 0.68 |
| 197 | TH-M-200095 | XIX | 0.64 |
| 564 | VGlut-F-000535 | XIX | 0.60 |
| 204 | Trh-M-100025 | XIV | 0.59 |
| 1003 | VGlut-F-000024 | XIX | 0.57 |
| 143 | Trh-M-400111 | XIX | 0.56 |
| 190 | TH-M-200014 | XIX | 0.55 |
| 1004 | VGlut-F-100016 | VI | 0.52 |
Cluster composition
This cluster has 22 neurons. The exemplar of this cluster (TH-F-100064) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-F-100064 | 13604 | THMARCM-544F_seg1 | TH-Gal4 | F | 67321.74 | 1.00 |
| TH-M-200037 | 3210 | THMARCM-333M_seg1 | TH-Gal4 | M | 47778.17 | 0.72 |
| TH-M-300045 | 3270 | THMARCM-465M_seg1 | TH-Gal4 | M | 47832.67 | 0.72 |
| TH-M-300052 | 3288 | THMARCM-485M_seg1 | TH-Gal4 | M | 48368.41 | 0.72 |
| Cha-F-300154 | 7129 | ChaMARCM-F000598_seg001 | Cha-Gal4 | F | 35668.28 | 0.58 |
| VGlut-F-000472 | 10999 | DvGlutMARCM-F002886_seg003 | VGlut-Gal4 | F | 45321.68 | 0.70 |
| TH-F-200004 | 13402 | THMARCM-194F_seg1 | TH-Gal4 | F | 49540.24 | 0.74 |
| TH-F-300035 | 13436 | THMARCM-251F_seg1 | TH-Gal4 | F | 49052.86 | 0.73 |
| TH-F-100033 | 13451 | THMARCM-290F_seg1 | TH-Gal4 | F | 49687.66 | 0.74 |
| TH-F-200068 | 13517 | THMARCM-393F_seg1 | TH-Gal4 | F | 46905.09 | 0.72 |
| TH-F-100063 | 13603 | THMARCM-542F_seg1 | TH-Gal4 | F | 47454.56 | 0.72 |
| TH-F-100068 | 13611 | THMARCM-555F_seg1 | TH-Gal4 | F | 47412.82 | 0.72 |
| TH-F-000036 | 13636 | THMARCM-623F_seg | TH-Gal4 | F | 49671.43 | 0.73 |
| TH-F-100075 | 13638 | THMARCM-632F_seg | TH-Gal4 | F | 49116.08 | 0.73 |
| TH-F-000057 | 13670 | THMARCM-695F-X2_seg1 | TH-Gal4 | F | 45077.45 | 0.69 |
| TH-F-000078 | 13733 | THMARCM-762F_seg1 | TH-Gal4 | F | 49381.46 | 0.74 |
| TH-F-000090 | 13758 | THMARCM-795F_seg2 | TH-Gal4 | F | 46568.06 | 0.72 |
| TH-F-100002 | 13780 | THMARCM-F_017_seg2 | TH-Gal4 | F | 43172.53 | 0.69 |
| VGlut-F-000268 | 14350 | DvGlutMARCM-F1034_seg1 | VGlut-Gal4 | F | 23297.49 | 0.33 |
| VGlut-F-500002 | 15596 | DvGlutMARCM-F012_seg1 | VGlut-Gal4 | F | 47700.02 | 0.69 |
| VGlut-F-000023 | 15779 | DvGlutMARCM-F185_seg1 | VGlut-Gal4 | F | 45264.59 | 0.71 |
| VGlut-F-000172 | 16201 | DvGlutMARCM-F582-X2_seg1 | VGlut-Gal4 | F | 48923.76 | 0.71 |