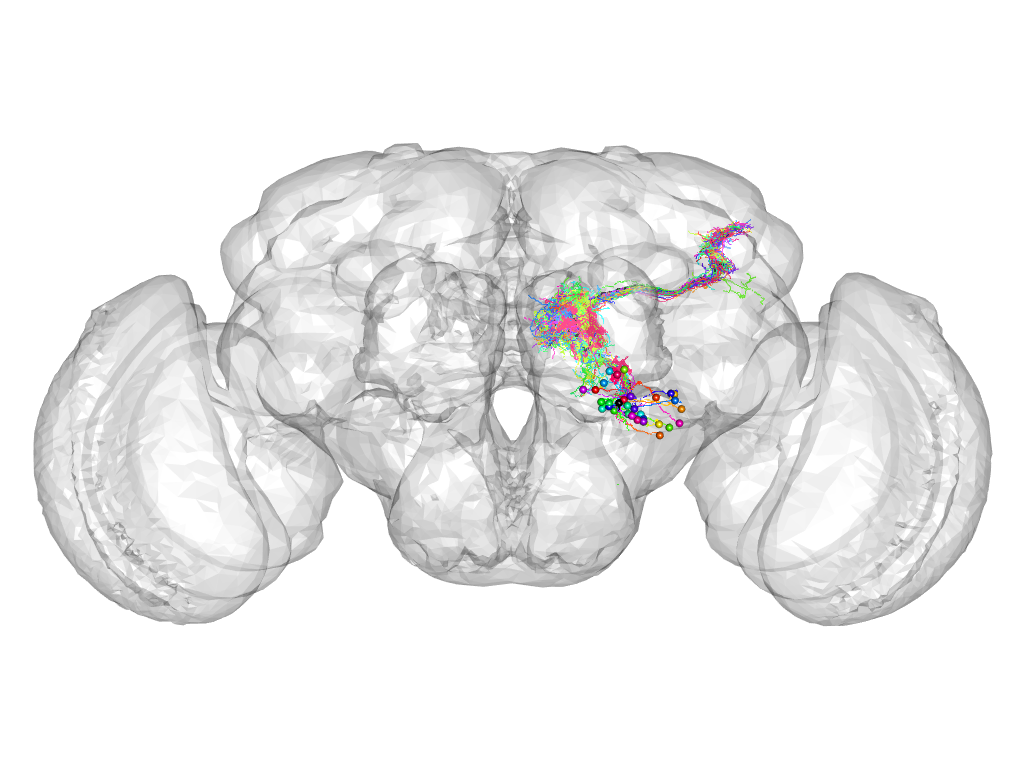
Cluster 799
Part of supercluster XI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-500124), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 911 | VGlut-F-400172 | XI | 0.74 |
| 842 | Trh-F-400067 | XI | 0.68 |
| 814 | VGlut-F-500300 | XI | 0.59 |
| 662 | Tdc2-F-400026 | XIII | 0.55 |
| 375 | VGlut-F-500848 | XI | 0.51 |
Cluster composition
This cluster has 33 neurons. The exemplar of this cluster (Trh-F-500124) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-500124 | 12456 | TPHMARCM-1247F_seg2 | Trh-Gal4 | F | 8496.37 | 1.00 |
| Trh-M-600076 | 1996 | TPHMARCM-M001493_seg001 | Trh-Gal4 | M | 5552.51 | 0.67 |
| Trh-M-500094 | 2661 | TPHMARCM-M001427_seg001 | Trh-Gal4 | M | 5747.18 | 0.66 |
| Trh-M-500146 | 2743 | TPHMARCM-M001509_seg001 | Trh-Gal4 | M | 6399.11 | 0.71 |
| Trh-M-400074 | 2845 | TPHMARCM-1071M_seg1 | Trh-Gal4 | M | 5975.88 | 0.67 |
| Trh-M-400082 | 2847 | TPHMARCM-1074M_seg1 | Trh-Gal4 | M | 6553.89 | 0.72 |
| Trh-M-700069 | 2876 | TPHMARCM-1121M_seg1 | Trh-Gal4 | M | 6376.76 | 0.70 |
| Trh-M-700024 | 3029 | TPHMARCM-981M_seg1 | Trh-Gal4 | M | 5300.65 | 0.61 |
| Trh-M-700025 | 3033 | TPHMARCM-986M_seg1 | Trh-Gal4 | M | 6115.84 | 0.62 |
| Trh-M-500018 | 3492 | TPHMARCM-363M_seg2 | Trh-Gal4 | M | 6312.40 | 0.70 |
| VGlut-F-700552 | 6651 | DvGlutMARCM-F004394_seg003 | VGlut-Gal4 | F | 4701.25 | 0.50 |
| VGlut-F-900119 | 6751 | DvGlutMARCM-F003956_seg001 | VGlut-Gal4 | F | 6272.32 | 0.71 |
| VGlut-F-800257 | 6971 | DvGlutMARCM-F004117_seg002 | VGlut-Gal4 | F | 5843.81 | 0.62 |
| VGlut-F-600691 | 7404 | DvGlutMARCM-F003862_seg001 | VGlut-Gal4 | F | 6043.42 | 0.66 |
| VGlut-F-800128 | 8458 | DvGlutMARCM-F003056_seg002 | VGlut-Gal4 | F | 5131.87 | 0.59 |
| Trh-F-500213 | 10455 | TPHMARCM-F001378_seg002 | Trh-Gal4 | F | 6293.03 | 0.73 |
| Trh-F-500220 | 10459 | TPHMARCM-F001518_seg003 | Trh-Gal4 | F | 6211.15 | 0.69 |
| VGlut-F-700243 | 11187 | DvGlutMARCM-F003035_seg001 | VGlut-Gal4 | F | 5566.64 | 0.60 |
| VGlut-F-400408 | 12328 | DvGlutMARCM-F1676_seg1 | VGlut-Gal4 | F | 5911.05 | 0.68 |
| Trh-F-500200 | 12573 | TPHMARCM-F001375_seg001 | Trh-Gal4 | F | 6440.62 | 0.71 |
| Trh-F-500211 | 12584 | TPHMARCM-F001377_seg001 | Trh-Gal4 | F | 6460.26 | 0.67 |
| Trh-F-500217 | 12608 | TPHMARCM-F001403_seg001 | Trh-Gal4 | F | 6517.60 | 0.75 |
| VGlut-F-700044 | 13070 | DvGlutMARCM-F2093_seg1 | VGlut-Gal4 | F | 6402.99 | 0.73 |
| Trh-F-700021 | 13243 | TPHMARCM-730F_seg1 | Trh-Gal4 | F | 6207.03 | 0.68 |
| Trh-F-500027 | 14020 | TPHMARCM-307F_seg1 | Trh-Gal4 | F | 6041.16 | 0.69 |
| Trh-F-500036 | 14072 | TPHMARCM-410F_seg1 | Trh-Gal4 | F | 6223.03 | 0.72 |
| Trh-F-600011 | 14100 | TPHMARCM-460F_seg1 | Trh-Gal4 | F | 6265.66 | 0.72 |
| Trh-F-700034 | 14270 | TPHMARCM-818F_seg1 | Trh-Gal4 | F | 6365.73 | 0.73 |
| Trh-F-700035 | 14271 | TPHMARCM-819F_seg1 | Trh-Gal4 | F | 5400.54 | 0.60 |
| VGlut-F-500197 | 15231 | DvGlutMARCM-F1553_seg2 | VGlut-Gal4 | F | 6255.45 | 0.67 |
| VGlut-F-400111 | 15489 | DvGlutMARCM-F646-X2_seg2 | VGlut-Gal4 | F | 6081.88 | 0.72 |
| VGlut-F-500038 | 15801 | DvGlutMARCM-F212_seg1 | VGlut-Gal4 | F | 6379.95 | 0.64 |
| VGlut-F-500039 | 15802 | DvGlutMARCM-F212_seg2 | VGlut-Gal4 | F | 6109.05 | 0.69 |