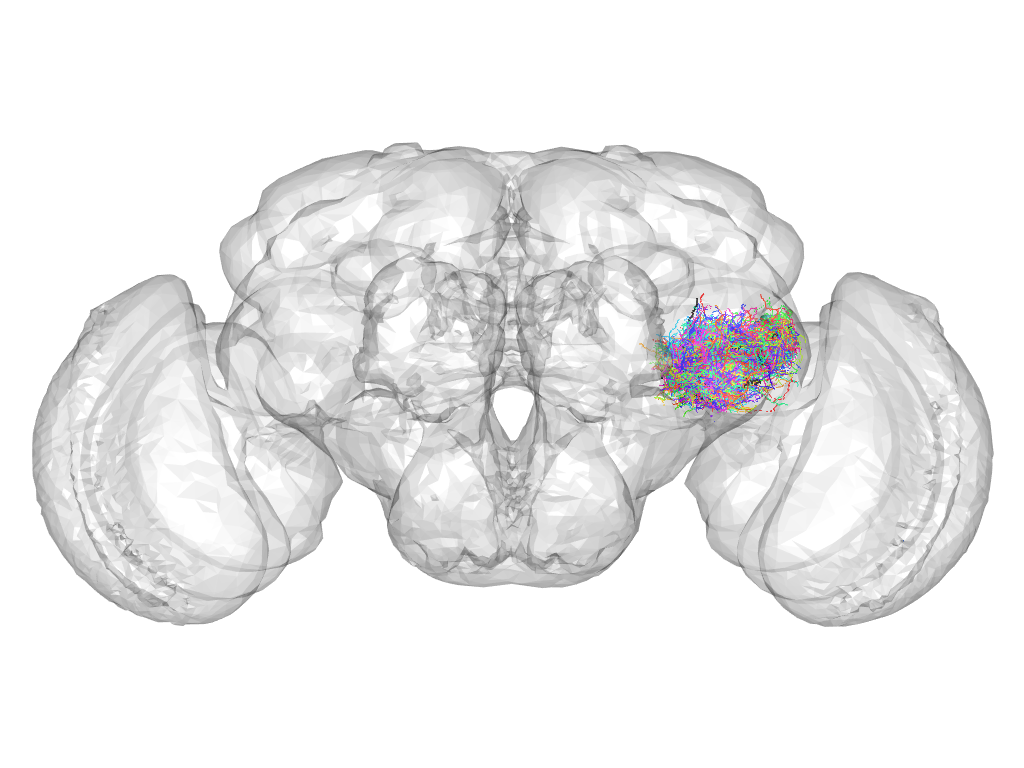
Cluster 641
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-600033), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 171 | Trh-M-000046 | X | 0.64 |
| 846 | Trh-F-700028 | X | 0.57 |
| 200 | TH-M-000060 | X | 0.56 |
| 876 | Trh-F-100023 | X | 0.52 |
| 180 | Trh-M-900013 | X | 0.49 |
Cluster composition
This cluster has 71 neurons. The exemplar of this cluster (fru-F-600033) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-600033 | 9988 | FruMARCM-F000577_seg001 | fru-Gal4 | F | 13849.30 | 1.00 |
| fru-M-700173 | 207 | FruMARCM-M002422_seg001 | fru-Gal4 | M | 9195.19 | 0.64 |
| fru-M-500151 | 447 | FruMARCM-M001558_seg001 | fru-Gal4 | M | 8697.05 | 0.56 |
| fru-M-700058 | 1365 | FruMARCM-M000875_seg001 | fru-Gal4 | M | 9462.87 | 0.67 |
| fru-M-500024 | 1944 | FruMARCM-M000540_seg001 | fru-Gal4 | M | 9133.53 | 0.67 |
| Trh-M-900030 | 2037 | TPHMARCM-M001549_seg001 | Trh-Gal4 | M | 8880.80 | 0.64 |
| Trh-M-600100 | 2200 | TPHMARCM-M001690_seg001 | Trh-Gal4 | M | 9307.02 | 0.68 |
| Trh-M-600113 | 2219 | TPHMARCM-M001710_seg001 | Trh-Gal4 | M | 9616.09 | 0.67 |
| Trh-M-600115 | 2221 | TPHMARCM-M001711_seg001 | Trh-Gal4 | M | 9538.15 | 0.69 |
| Trh-M-600116 | 2222 | TPHMARCM-M001712_seg001 | Trh-Gal4 | M | 9391.43 | 0.65 |
| Trh-M-000107 | 2293 | TPHMARCM-M001775_seg001 | Trh-Gal4 | M | 9136.10 | 0.64 |
| Trh-M-500169 | 2558 | TPHMARCM-M001535_seg001 | Trh-Gal4 | M | 9436.96 | 0.67 |
| Trh-M-600069 | 2612 | TPHMARCM-1318M_seg1 | Trh-Gal4 | M | 9161.63 | 0.68 |
| Trh-M-500097 | 2664 | TPHMARCM-M001428_seg001 | Trh-Gal4 | M | 9224.97 | 0.68 |
| Trh-M-500112 | 2700 | TPHMARCM-M001464_seg003 | Trh-Gal4 | M | 8821.47 | 0.65 |
| Trh-M-400105 | 2746 | TPHMARCM-M001512_seg001 | Trh-Gal4 | M | 9142.87 | 0.68 |
| Trh-M-500148 | 2753 | TPHMARCM-M001520_seg001 | Trh-Gal4 | M | 9540.60 | 0.64 |
| Trh-M-700046 | 2758 | TPHMARCM-1022M_seg1 | Trh-Gal4 | M | 9142.53 | 0.65 |
| Trh-M-700047 | 2759 | TPHMARCM-1022M_seg2 | Trh-Gal4 | M | 8843.06 | 0.63 |
| Trh-M-700038 | 2781 | TPHMARCM-1000M_seg1 | Trh-Gal4 | M | 8770.49 | 0.63 |
| Trh-M-500081 | 2820 | TPHMARCM-1049M_seg1 | Trh-Gal4 | M | 8320.82 | 0.63 |
| Trh-M-500089 | 2872 | TPHMARCM-1115M_seg1 | Trh-Gal4 | M | 9310.45 | 0.69 |
| Trh-M-900017 | 2951 | TPHMARCM-892M_seg3 | Trh-Gal4 | M | 9011.12 | 0.65 |
| Trh-M-900022 | 2983 | TPHMARCM-920M_seg1 | Trh-Gal4 | M | 9052.82 | 0.65 |
| Trh-M-800017 | 2990 | TPHMARCM-932M_seg1 | Trh-Gal4 | M | 8485.32 | 0.63 |
| Trh-M-500077 | 3030 | TPHMARCM-983M_seg1 | Trh-Gal4 | M | 8277.54 | 0.58 |
| Trh-M-700026 | 3034 | TPHMARCM-987M_seg1 | Trh-Gal4 | M | 7713.45 | 0.61 |
| Trh-M-500002 | 3434 | TPHMARCM-181M_seg1 | Trh-Gal4 | M | 9363.65 | 0.68 |
| Trh-M-500009 | 3446 | TPHMARCM-195M_seg1 | Trh-Gal4 | M | 9022.66 | 0.65 |
| Trh-M-400026 | 3613 | TPHMARCM-573M_seg1 | Trh-Gal4 | M | 9384.06 | 0.67 |
| fru-F-500224 | 4670 | FruMARCM-F001830_seg001 | fru-Gal4 | F | 9410.78 | 0.63 |
| fru-F-200142 | 4691 | FruMARCM-F001924_seg002 | fru-Gal4 | F | 9208.78 | 0.67 |
| fru-F-800058 | 4701 | FruMARCM-F001961_seg001 | fru-Gal4 | F | 9542.87 | 0.70 |
| fru-F-500239 | 4730 | FruMARCM-F002057_seg001 | fru-Gal4 | F | 9720.00 | 0.65 |
| fru-F-500255 | 4784 | FruMARCM-F002105_seg001 | fru-Gal4 | F | 9400.28 | 0.67 |
| Cha-F-100081 | 5259 | ChaMARCM-F000657_seg002 | Cha-Gal4 | F | 8984.22 | 0.66 |
| Cha-F-100094 | 5453 | ChaMARCM-F000790_seg001 | Cha-Gal4 | F | 9065.72 | 0.65 |
| VGlut-F-800316 | 5912 | DvGlutMARCM-F004577_seg002 | VGlut-Gal4 | F | 9549.82 | 0.69 |
| VGlut-F-600156 | 6106 | DvGlutMARCM-F2171_seg2 | VGlut-Gal4 | F | 9235.24 | 0.66 |
| VGlut-F-400816 | 6737 | DvGlutMARCM-F003942_seg001 | VGlut-Gal4 | F | 9292.55 | 0.66 |
| Cha-F-500125 | 7091 | ChaMARCM-F000569_seg002 | Cha-Gal4 | F | 8726.71 | 0.64 |
| fru-F-500109 | 7763 | FruMARCM-F000662_seg002 | fru-Gal4 | F | 9177.28 | 0.68 |
| fru-F-500120 | 7793 | FruMARCM-F000688_seg003 | fru-Gal4 | F | 9772.89 | 0.69 |
| VGlut-F-500568 | 8556 | DvGlutMARCM-F003124_seg002 | VGlut-Gal4 | F | 9368.98 | 0.69 |
| VGlut-F-700302 | 8575 | DvGlutMARCM-F003141_seg001 | VGlut-Gal4 | F | 9119.18 | 0.62 |
| fru-F-700047 | 9950 | FruMARCM-F000543_seg001 | fru-Gal4 | F | 9222.78 | 0.67 |
| fru-F-500085 | 10017 | FruMARCM-F000604_seg004 | fru-Gal4 | F | 9421.06 | 0.67 |
| VGlut-F-700196 | 10932 | DvGlutMARCM-F002827_seg002 | VGlut-Gal4 | F | 8662.59 | 0.60 |
| VGlut-F-600470 | 11093 | DvGlutMARCM-F002966_seg002 | VGlut-Gal4 | F | 9353.99 | 0.67 |
| fru-F-800003 | 11444 | FruMARCM-F000064_seg001 | fru-Gal4 | F | 9845.34 | 0.68 |
| VGlut-F-800069 | 12121 | DvGlutMARCM-F002617_seg002 | VGlut-Gal4 | F | 9794.78 | 0.65 |
| Trh-F-500117 | 12416 | TPHMARCM-1209F_seg1 | Trh-Gal4 | F | 8883.59 | 0.58 |
| Trh-F-500171 | 12521 | TPHMARCM-1303F_seg1 | Trh-Gal4 | F | 8753.25 | 0.65 |
| Trh-F-500173 | 12529 | TPHMARCM-1311F_seg1 | Trh-Gal4 | F | 8766.96 | 0.64 |
| Trh-F-500175 | 12531 | TPHMARCM-1311F_seg3 | Trh-Gal4 | F | 9744.18 | 0.71 |
| Trh-F-500184 | 12542 | TPHMARCM-1330F_seg1 | Trh-Gal4 | F | 9514.37 | 0.70 |
| Trh-F-500195 | 12563 | TPHMARCM-F001351_seg001 | Trh-Gal4 | F | 8693.58 | 0.65 |
| Trh-F-900034 | 12736 | TPHMARCM-958F_seg1 | Trh-Gal4 | F | 9161.45 | 0.70 |
| Trh-F-900035 | 12737 | TPHMARCM-959F_seg1 | Trh-Gal4 | F | 8818.61 | 0.65 |
| Trh-F-500008 | 13855 | TPHMARCM-125F_seg3 | Trh-Gal4 | F | 9885.43 | 0.68 |
| Trh-F-500009 | 13856 | TPHMARCM-126F_seg1 | Trh-Gal4 | F | 9458.11 | 0.64 |
| Trh-F-500025 | 14016 | TPHMARCM-303F_seg1 | Trh-Gal4 | F | 9313.07 | 0.66 |
| Trh-F-500026 | 14017 | TPHMARCM-304F_seg1 | Trh-Gal4 | F | 8985.01 | 0.65 |
| Trh-F-600008 | 14097 | TPHMARCM-458F_seg1 | Trh-Gal4 | F | 9076.93 | 0.65 |
| Trh-F-500044 | 14119 | TPHMARCM-479F_seg1 | Trh-Gal4 | F | 9084.62 | 0.63 |
| Trh-F-600032 | 14204 | TPHMARCM-645F_seg1 | Trh-Gal4 | F | 9161.10 | 0.68 |
| Trh-F-700014 | 14226 | TPHMARCM-679F_seg1 | Trh-Gal4 | F | 9409.83 | 0.69 |
| Trh-F-700015 | 14227 | TPHMARCM-679F_seg2 | Trh-Gal4 | F | 9203.23 | 0.67 |
| VGlut-F-400228 | 14537 | DvGlutMARCM-F806-x2_seg2 | VGlut-Gal4 | F | 9130.26 | 0.67 |
| VGlut-F-500138 | 14661 | DvGlutMARCM-F922_seg1 | VGlut-Gal4 | F | 9614.17 | 0.69 |
| VGlut-F-000351 | 15041 | DvGlutMARCM-F1379_seg1 | VGlut-Gal4 | F | 9197.45 | 0.69 |