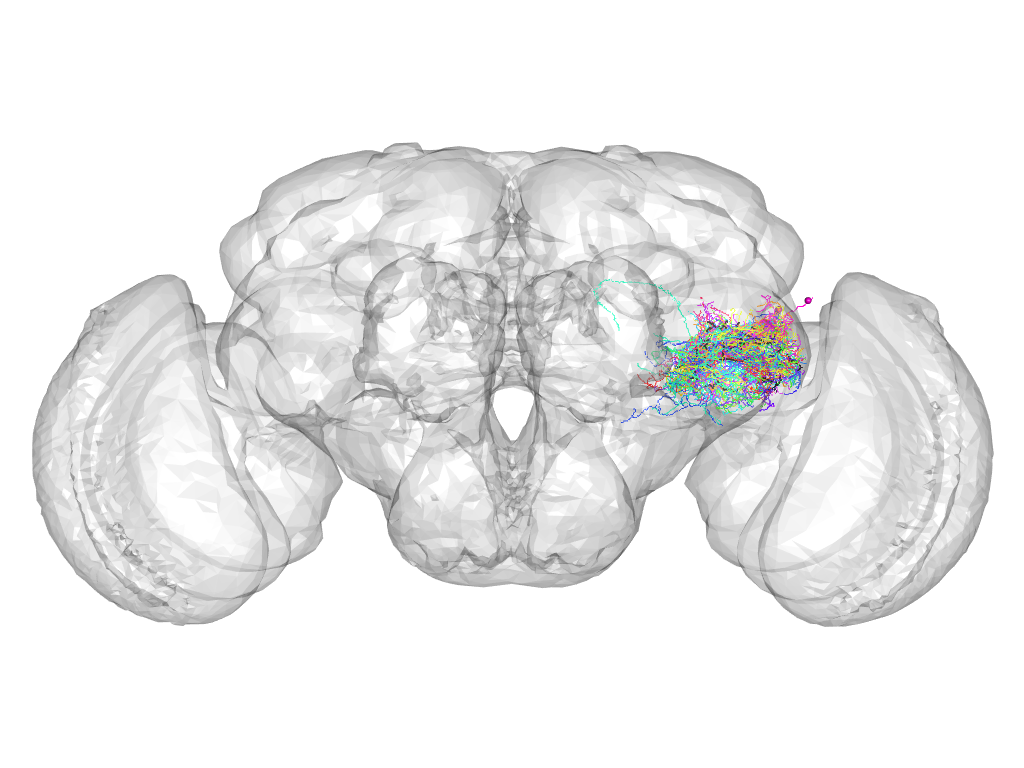
Cluster 846
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-700028), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 180 | Trh-M-900013 | X | 0.70 |
| 321 | Cha-F-600151 | X | 0.66 |
| 641 | fru-F-600033 | X | 0.61 |
| 145 | Trh-M-200079 | VII | 0.58 |
| 171 | Trh-M-000046 | X | 0.57 |
| 939 | VGlut-F-400341 | X | 0.52 |
Cluster composition
This cluster has 23 neurons. The exemplar of this cluster (Trh-F-700028) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-700028 | 13306 | TPHMARCM-788F_seg1 | Trh-Gal4 | F | 9475.84 | 1.00 |
| Trh-M-400076 | 922 | TPHMARCM-1072M_seg001 | Trh-Gal4 | M | 6423.13 | 0.65 |
| fru-M-500079 | 1393 | FruMARCM-M000895_seg001 | fru-Gal4 | M | 6125.69 | 0.66 |
| fru-M-400073 | 1424 | FruMARCM-M000937_seg001 | fru-Gal4 | M | 6269.05 | 0.61 |
| Trh-M-600035 | 1986 | TPHMARCM-0770M_seg001 | Trh-Gal4 | M | 6338.41 | 0.66 |
| Trh-M-500099 | 2678 | TPHMARCM-M001444_seg001 | Trh-Gal4 | M | 6170.95 | 0.59 |
| Trh-M-800011 | 2958 | TPHMARCM-899M_seg1 | Trh-Gal4 | M | 6334.94 | 0.65 |
| Trh-M-800019 | 2991 | TPHMARCM-934M_seg1 | Trh-Gal4 | M | 5940.76 | 0.65 |
| Trh-M-600055 | 3032 | TPHMARCM-985M_seg1 | Trh-Gal4 | M | 6564.03 | 0.65 |
| fru-F-500198 | 4529 | FruMARCM-F001640_seg001 | fru-Gal4 | F | 6712.01 | 0.69 |
| fru-F-100067 | 4769 | FruMARCM-F002088_seg001 | fru-Gal4 | F | 6688.05 | 0.68 |
| Cha-F-000216 | 5021 | ChaMARCM-F001109_seg003 | Cha-Gal4 | F | 5497.91 | 0.45 |
| fru-F-400130 | 6186 | FruMARCM-F001118_seg001 | fru-Gal4 | F | 6367.63 | 0.65 |
| fru-F-700128 | 6256 | FruMARCM-F001286_seg001 | fru-Gal4 | F | 6780.79 | 0.68 |
| VGlut-F-900122 | 6461 | DvGlutMARCM-F004246_seg001 | VGlut-Gal4 | F | 6458.94 | 0.66 |
| Cha-F-500060 | 8177 | ChaMARCM-F000194_seg002 | Cha-Gal4 | F | 5348.67 | 0.49 |
| fru-F-600011 | 9918 | FruMARCM-F000501_seg001 | fru-Gal4 | F | 6857.05 | 0.70 |
| Trh-F-500134 | 12469 | TPHMARCM-1258F_seg1 | Trh-Gal4 | F | 6600.69 | 0.67 |
| Trh-F-500180 | 12538 | TPHMARCM-1321F_seg1 | Trh-Gal4 | F | 6117.35 | 0.64 |
| Trh-F-600038 | 13263 | TPHMARCM-751F_seg1 | Trh-Gal4 | F | 6539.17 | 0.64 |
| Trh-F-900019 | 13275 | TPHMARCM-761F_seg1 | Trh-Gal4 | F | 5850.75 | 0.54 |
| Trh-F-700029 | 13307 | TPHMARCM-789F_seg1 | Trh-Gal4 | F | 6098.30 | 0.65 |
| Trh-F-400013 | 13868 | TPHMARCM-138F_seg1 | Trh-Gal4 | F | 6687.29 | 0.68 |